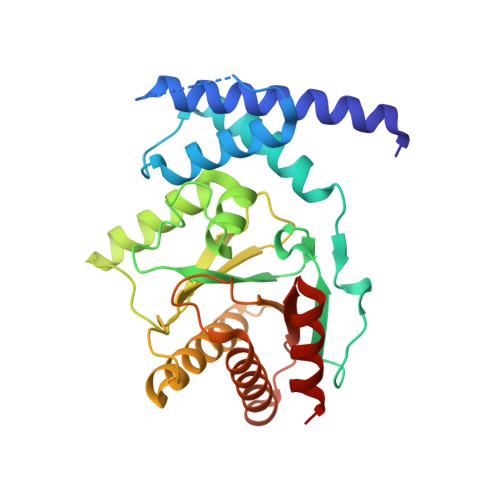
Structure of the sirtuin-linked macrodomain SAV0325 from Staphylococcus aureus.
Appel, C.D., Feld, G.K., Wallace, B.D., Williams, R.S.(2016) Protein Sci 25: 1682-1691
- PubMed: 27345688
- DOI: https://doi.org/10.1002/pro.2974
- Primary Citation of Related Structures:
5KIV - PubMed Abstract:
Cells use the post-translational modification ADP-ribosylation to control a host of biological activities. In some pathogenic bacteria, an operon-encoded mono-ADP-ribosylation cycle mediates response to host-induced oxidative stress. In this system, reversible mono ADP-ribosylation of a lipoylated target protein represses oxidative stress response. An NAD(+) -dependent sirtuin catalyzes the single ADP-ribose (ADPr) addition, while a linked macrodomain-containing protein removes the ADPr. Here we report the crystal structure of the sitruin-linked macrodomain protein from Staphylococcus aureus, SauMacro (also known as SAV0325) to 1.75-Å resolution. The monomeric SauMacro bears a previously unidentified Zn(2+) -binding site that putatively aids in substrate recognition and catalysis. An amino-terminal three-helix bundle motif unique to this class of macrodomain proteins provides a structural scaffold for the Zn(2+) site. Structural features of the enzyme further indicate a cleft proximal to the Zn(2+) binding site appears well suited for ADPr binding, while a deep hydrophobic channel in the protein core is suitable for binding the lipoate of the lipoylated protein target.
Organizational Affiliation:
Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, US National Institutes of Health, Department of Health and Human Services, Research Triangle Park, North Carolina, 27709.