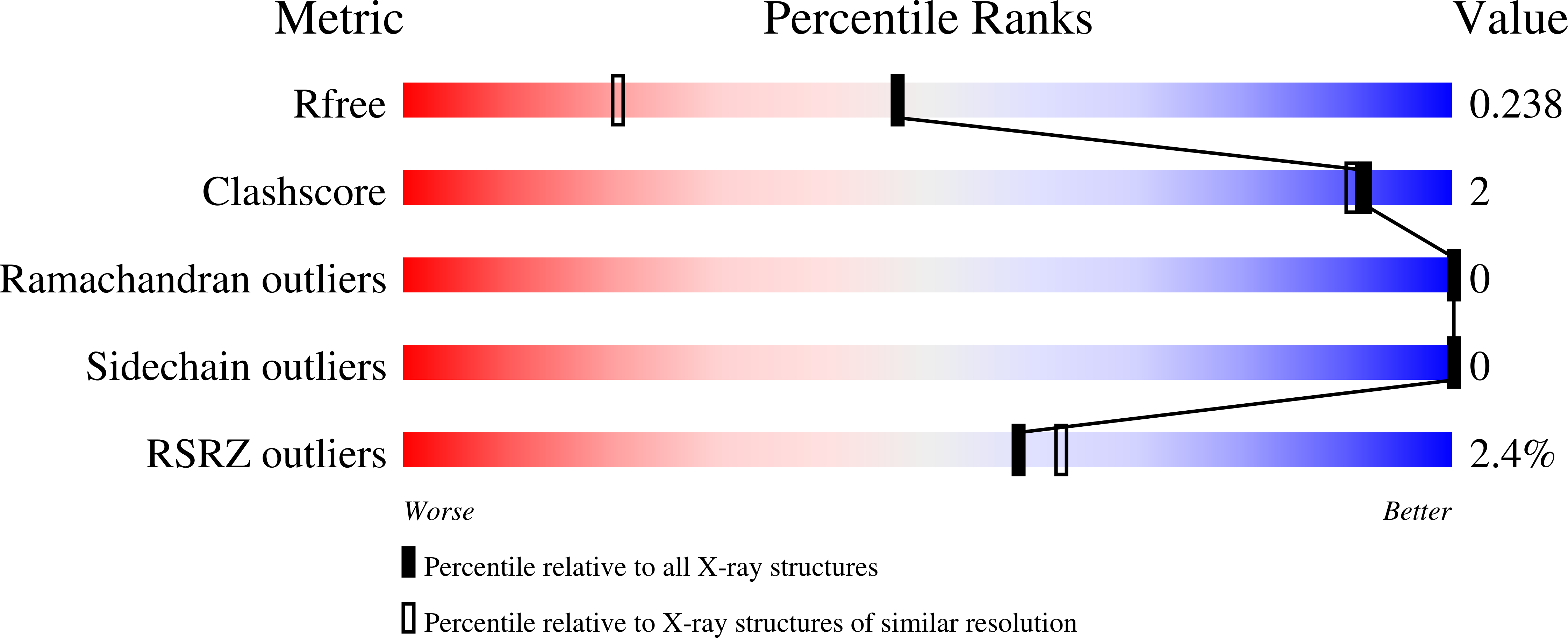
Modular Artificial Cupredoxins.
Mann, S.I., Heinisch, T., Weitz, A.C., Hendrich, M.P., Ward, T.R., Borovik, A.S.(2016) J Am Chem Soc 138: 9073-9076
- PubMed: 27385206
- DOI: https://doi.org/10.1021/jacs.6b05428
- Primary Citation of Related Structures:
5K67, 5K68, 5L3Y, 5WBC - PubMed Abstract:
Cupredoxins are electron-transfer proteins that have active sites containing a mononuclear Cu center with an unusual trigonal monopyramidal structure (Type 1 Cu). A single Cu-Scys bond is present within the trigonal plane that is responsible for its unique physical properties. We demonstrate that a cysteine-containing variant of streptavidin (Sav) can serve as a protein host to model the structure and properties of Type 1 Cu sites. A series of artificial Cu proteins are described that rely on Sav and a series of biotinylated synthetic Cu complexes. Optical and EPR measurements highlight the presence of a Cu-Scys bond, and XRD analysis provides structural evidence. We further provide evidence that changes in the linker between the biotin and Cu complex within the synthetic constructs allows for small changes in the placement of Cu centers within Sav that have dramatic effects on the structural and physical properties of the resulting artificial metalloproteins. These findings highlight the utility of the biotin-Sav technology as an approach for simulating active sites of metalloproteins.
Organizational Affiliation:
Department of Chemistry, University of California-Irvine , 1102 Natural Sciences II, Irvine, California 92697, United States.