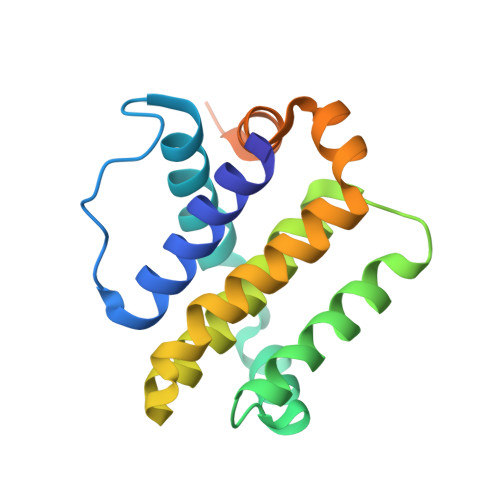
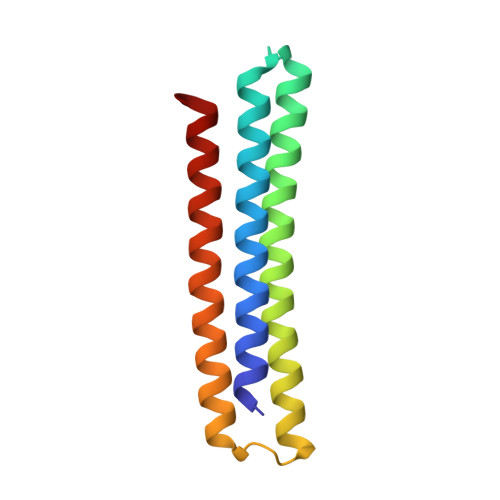
Computationally designed high specificity inhibitors delineate the roles of BCL2 family proteins in cancer.
Berger, S., Procko, E., Margineantu, D., Lee, E.F., Shen, B.W., Zelter, A., Silva, D.A., Chawla, K., Herold, M.J., Garnier, J.M., Johnson, R., MacCoss, M.J., Lessene, G., Davis, T.N., Stayton, P.S., Stoddard, B.L., Fairlie, W.D., Hockenbery, D.M., Baker, D.(2016) Elife 5
- PubMed: 27805565
- DOI: https://doi.org/10.7554/eLife.20352
- Primary Citation of Related Structures:
5JSB, 5JSN - PubMed Abstract:
Many cancers overexpress one or more of the six human pro-survival BCL2 family proteins to evade apoptosis. To determine which BCL2 protein or proteins block apoptosis in different cancers, we computationally designed three-helix bundle protein inhibitors specific for each BCL2 pro-survival protein. Following in vitro optimization, each inhibitor binds its target with high picomolar to low nanomolar affinity and at least 300-fold specificity. Expression of the designed inhibitors in human cancer cell lines revealed unique dependencies on BCL2 proteins for survival which could not be inferred from other BCL2 profiling methods. Our results show that designed inhibitors can be generated for each member of a closely-knit protein family to probe the importance of specific protein-protein interactions in complex biological processes.
Organizational Affiliation:
Department of Bioengineering, University of Washington, Seattle, United States.