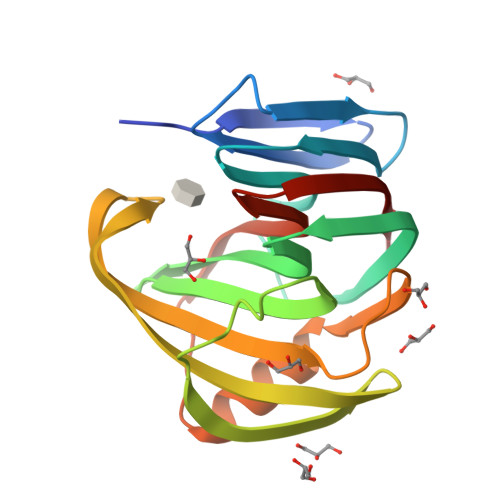
Structural and functional characterization of a highly stable endo-beta-1,4-xylanase from Fusarium oxysporum and its development as an efficient immobilized biocatalyst.
Gomez, S., Payne, A.M., Savko, M., Fox, G.C., Shepard, W.E., Fernandez, F.J., Cristina Vega, M.(2016) Biotechnol Biofuels 9: 191-191
- PubMed: 27602054
- DOI: https://doi.org/10.1186/s13068-016-0605-z
- Primary Citation of Related Structures:
5JRM, 5JRN - PubMed Abstract:
Replacing fossil fuel with renewable sources such as lignocellulosic biomass is currently a promising alternative for obtaining biofuel and for fighting against the consequences of climate change. However, the recalcitrant structure of lignocellulosic biomass residues constitutes a major limitation for its widespread use in industry. The efficient hydrolysis of lignocellulosic materials requires the complementary action of multiple enzymes including xylanases and β-xylosidases, which are responsible for cleaving exo- and endoxylan linkages, that release oligocarbohydrates that can be further processed by other enzymes. We have identified the endo-β-1,4-xylanase Xyl2 from Fusarium oxysporum as a promising glycoside hydrolase family 11 enzyme for the industrial degradation of xylan. To characterize Xyl2, we have cloned the synthetic optimized gene and expressed and purified recombinant Xyl2 to homogeneity, finally obtaining 10 mg pure Xyl2 per liter of culture. The crystal structure of Xyl2 at 1.56 Å resolution and the structure of a methyl-xylopyranoside Xyl2 complex at 2.84 Å resolution cast a highly detailed view of the active site of the enzyme, revealing the molecular basis for the high catalytic efficiency of Xyl2. The kinetic analysis of Xyl2 demonstrates high xylanase activity and non-negligible β-xylosidase activity under a variety of experimental conditions including alkaline pH and elevated temperature. Immobilizing Xyl2 on a variety of solid supports enhances the enzymatic properties that render Xyl2 a promising industrial biocatalyst, which, together with the detailed structural data, may establish Xyl2 as a platform for future developments of industrially relevant xylanases. F. oxysporum Xyl2 is a GH11 xylanase which is highly active in free form and immobilized onto a variety of solid supports in a wide pH range. Furthermore, immobilization of Xyl2 on certain supports significantly increases its thermal stability. A mechanistic rationale for Xyl2's remarkable catalytic efficiency at alkaline pH is proposed on the basis of two crystallographic structures. Together, these properties render Xyl2 an attractive biocatalyst for the sustainable industrial degradation of xylan.
Organizational Affiliation:
Structural and Quantitative Biology Department, Center for Biological Research (CIB-CSIC), Ramiro de Maeztu 9, 28040 Madrid, Spain.