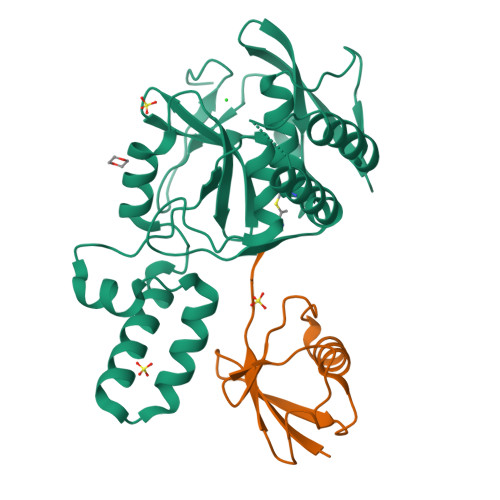
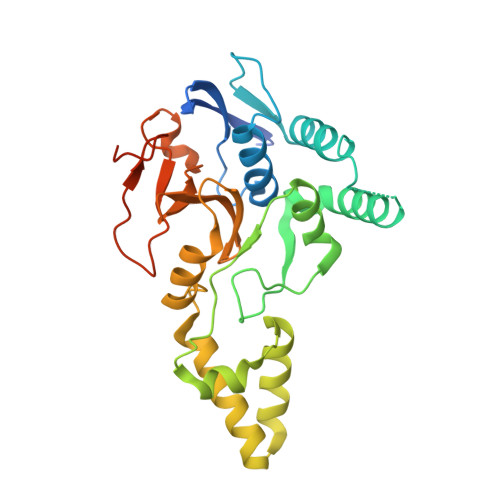
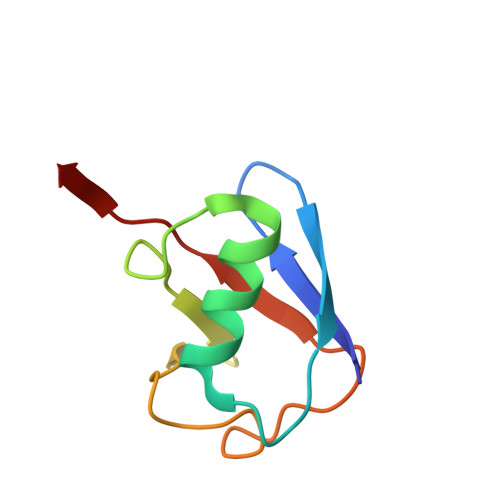
MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes.
Abdul Rehman, S.A., Kristariyanto, Y.A., Choi, S.Y., Nkosi, P.J., Weidlich, S., Labib, K., Hofmann, K., Kulathu, Y.(2016) Mol Cell 63: 146-155
- PubMed: 27292798
- DOI: https://doi.org/10.1016/j.molcel.2016.05.009
- Primary Citation of Related Structures:
5JKN, 5JQS - PubMed Abstract:
Deubiquitinating enzymes (DUBs) remove ubiquitin (Ub) from Ub-conjugated substrates to regulate the functional outcome of ubiquitylation. Here we report the discovery of a new family of DUBs, which we have named MINDY (motif interacting with Ub-containing novel DUB family). Found in all eukaryotes, MINDY-family DUBs are highly selective at cleaving K48-linked polyUb, a signal that targets proteins for degradation. We identify the catalytic activity to be encoded within a previously unannotated domain, the crystal structure of which reveals a distinct protein fold with no homology to any of the known DUBs. The crystal structure of MINDY-1 (also known as FAM63A) in complex with propargylated Ub reveals conformational changes that realign the active site for catalysis. MINDY-1 prefers cleaving long polyUb chains and works by trimming chains from the distal end. Collectively, our results reveal a new family of DUBs that may have specialized roles in regulating proteostasis.
Organizational Affiliation:
MRC Protein Phosphorylation & Ubiquitylation Unit, School of Life Sciences, University of Dundee, Dow Street, Dundee DD1 5EH, UK.