Structural and histone binding ability characterization of the ARB2 domain of a histone deacetylase Hda1 from Saccharomyces cerevisiae.
Shen, H., Zhu, Y., Wang, C., Yan, H., Teng, M., Li, X.(2016) Sci Rep 6: 33905-33905
- PubMed: 27665728
- DOI: https://doi.org/10.1038/srep33905
- Primary Citation of Related Structures:
5J8J - PubMed Abstract:
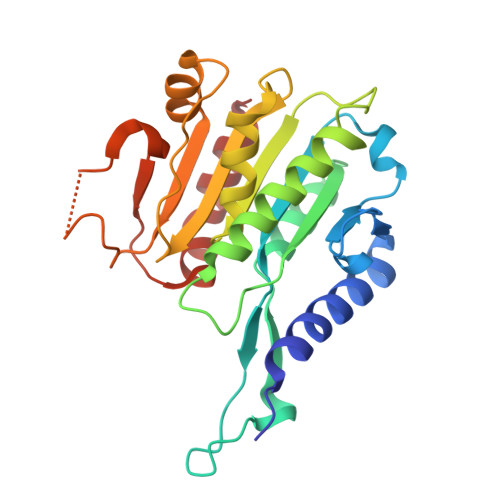
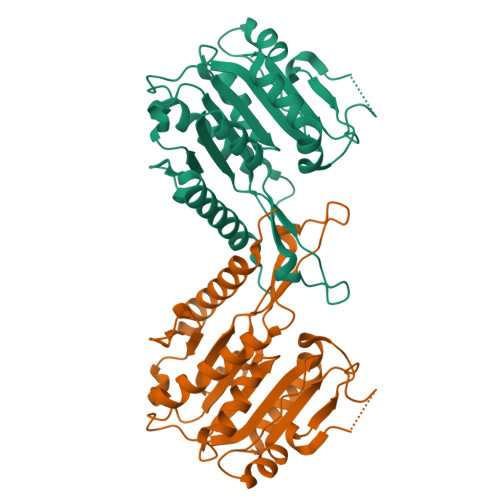
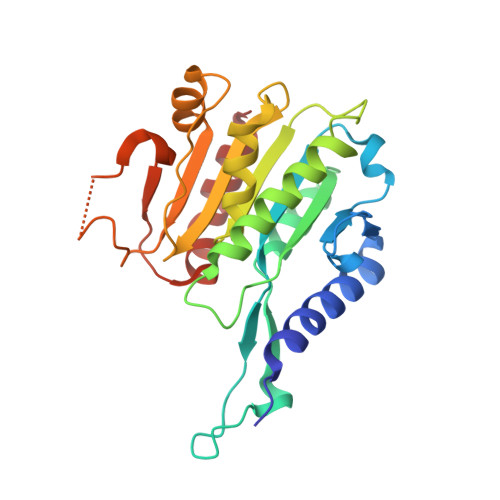
Hda1 is the catalytic core component of the H2B- and H3- specific histone deacetylase (HDAC) complex from Saccharomyces cerevisiae, which is involved in the epigenetic repression and plays a crucial role in transcriptional regulation and developmental events. Though the N-terminal catalytic HDAC domain of Hda1 is well characterized, the function of the C-terminal ARB2 domain remains unknown. In this study, we determine the crystal structure of the ARB2 domain from S. cerevisiae Hda1 at a resolution of 2.7 Å. The ARB2 domain displays an α/β sandwich architecture with an arm protruding outside. Two ARB2 domain molecules form a compact homo-dimer via the arm elements, and assemble as an inverse "V" shape. The pull-down and ITC results reveal that the ARB2 domain possesses the histone binding ability, recognizing both the H2A-H2B dimer and H3-H4 tetramer. Perturbation of the dimer interface abolishes the histone binding ability of the ARB2 domain, indicating that the unique dimer architecture of the ARB2 domain coincides with the function for anchoring to histone. Collectively, our data report the first structure of the ARB2 domain and disclose its histone binding ability, which is of benefit for understanding the deacetylation reaction catalyzed by the class II Hda1 HDAC complex.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale, Innovation Center for Cell Signaling Network, School of Life Science, University of Science and Technology of China, Hefei, Anhui, 230026, People's Republic of China.