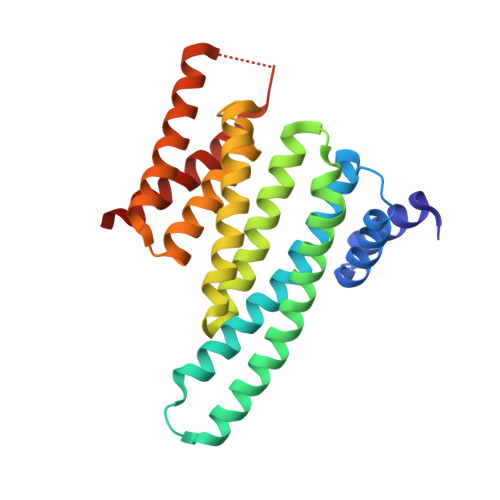
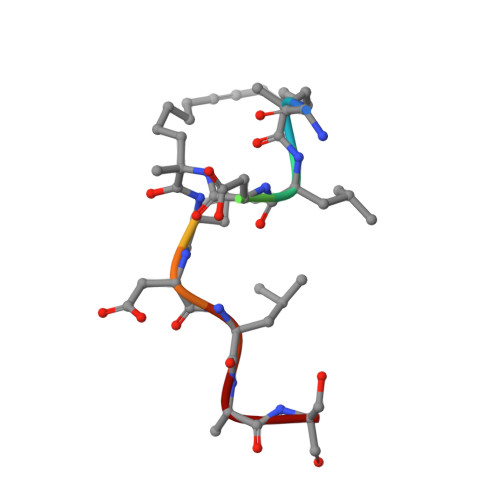
Constraining an Irregular Peptide Secondary Structure through Ring-Closing Alkyne Metathesis.
Cromm, P.M., Wallraven, K., Glas, A., Bier, D., Furstner, A., Ottmann, C., Grossmann, T.N.(2016) Chembiochem 17: 1915-1919
- PubMed: 27596722
- DOI: https://doi.org/10.1002/cbic.201600362
- Primary Citation of Related Structures:
5J31 - PubMed Abstract:
Macrocyclization can be used to constrain peptides in their bioactive conformations, thereby supporting target affinity and bioactivity. In particular, for the targeting of challenging protein-protein interactions, macrocyclic peptides have proven to be very useful. Available approaches focus on the stabilization of α-helices, which limits their general applicability. Here we report for the first time on the use of ring-closing alkyne metathesis for the stabilization of an irregular peptide secondary structure. A small library of alkyne-crosslinked peptides provided a number of derivatives with improved target affinity relative to the linear parent peptide. In addition, we report the crystal structure of the highest-affinity derivative in a complex with its protein target 14-3-3ζ. It can be expected that the alkyne-based macrocyclization of irregular binding epitopes should give rise to new scaffolds suitable for targeting of currently intractable proteins.
Organizational Affiliation:
Department of Chemical Biology, Max-Planck-Institute of Molecular Physiology, Otto-Hahn-Strasse 11, 44227, Dortmund, Germany.