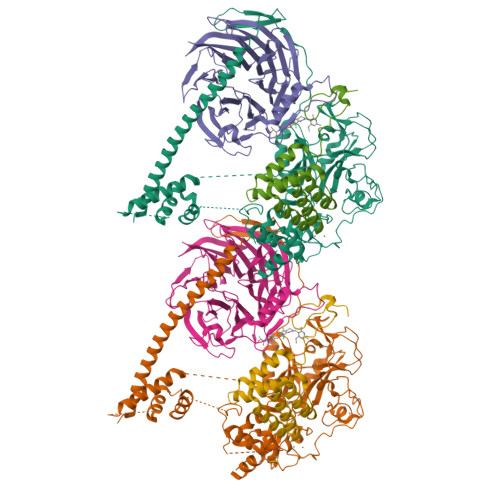
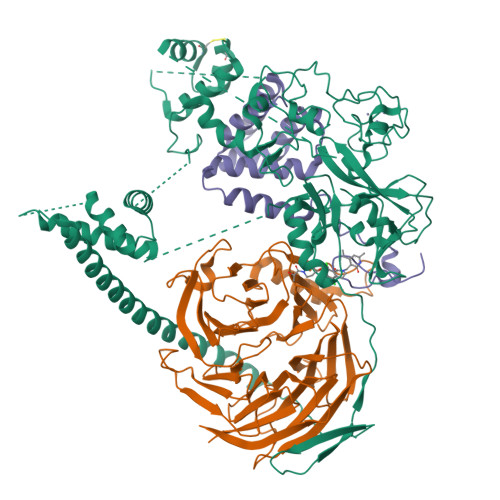
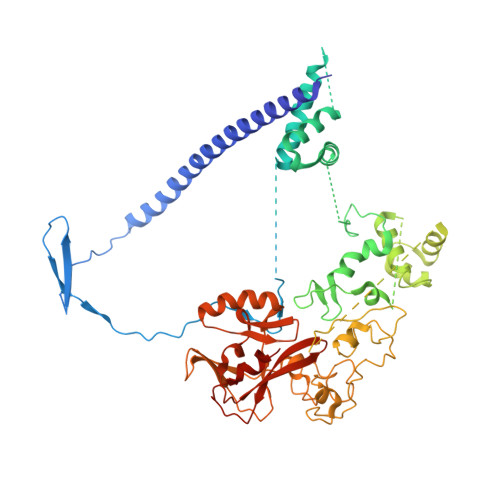
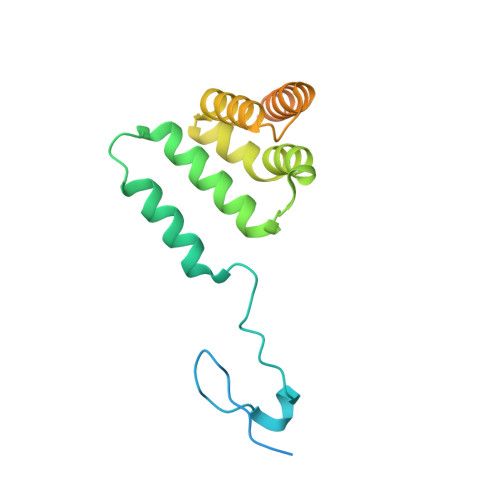
Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance.
Brooun, A., Gajiwala, K.S., Deng, Y.L., Liu, W., Bolanos, B., Bingham, P., He, Y.A., Diehl, W., Grable, N., Kung, P.P., Sutton, S., Maegley, K.A., Yu, X., Stewart, A.E.(2016) Nat Commun 7: 11384-11384
- PubMed: 27122193
- DOI: https://doi.org/10.1038/ncomms11384
- Primary Citation of Related Structures:
5IJ7, 5IJ8 - PubMed Abstract:
Polycomb repressive complex 2 (PRC2) mediates gene silencing through chromatin reorganization by methylation of histone H3 lysine 27 (H3K27). Overexpression of the complex and point mutations in the individual subunits of PRC2 have been shown to contribute to tumorigenesis. Several inhibitors of the PRC2 activity have shown efficacy in EZH2-mutated lymphomas and are currently in clinical development, although the molecular basis of inhibitor recognition remains unknown. Here we report the crystal structures of the inhibitor-bound wild-type and Y641N PRC2. The structures illuminate an important role played by a stretch of 17 residues in the N-terminal region of EZH2, we call the activation loop, in the stimulation of the enzyme activity, inhibitor recognition and the potential development of the mutation-mediated drug resistance. The work presented here provides new avenues for the design and development of next-generation PRC2 inhibitors through establishment of a structure-based drug design platform.
Organizational Affiliation:
Worldwide Medicinal Chemistry, Worldwide Research and Development, Pfizer Inc., San Diego, California 92121, USA.