A crystallographic study of human NONO (p54(nrb)): overcoming pathological problems with purification, data collection and noncrystallographic symmetry.
Knott, G.J., Panjikar, S., Thorn, A., Fox, A.H., Conte, M.R., Lee, M., Bond, C.S.(2016) Acta Crystallogr D Struct Biol 72: 761-769
- PubMed: 27303796
- DOI: https://doi.org/10.1107/S2059798316005830
- Primary Citation of Related Structures:
5IFM - PubMed Abstract:
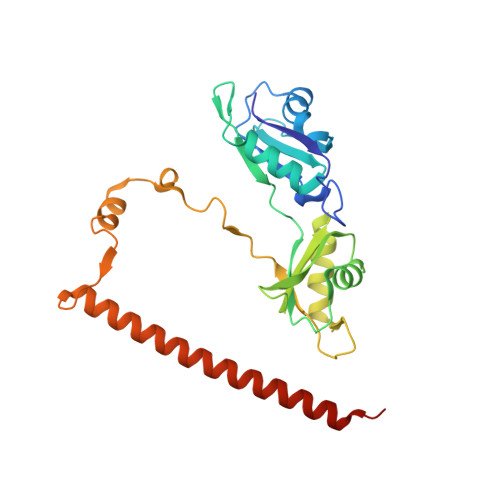
Non-POU domain-containing octamer-binding protein (NONO, a.k.a. p54(nrb)) is a central player in nuclear gene regulation with rapidly emerging medical significance. NONO is a member of the highly conserved Drosophila behaviour/human splicing (DBHS) protein family, a dynamic family of obligatory dimeric nuclear regulatory mediators. However, work with the NONO homodimer has been limited by rapid irreversible sample aggregation. Here, it is reported that L-proline stabilizes purified NONO homodimers, enabling good-quality solution small-angle X-ray structure determination and crystallization. NONO crystallized in the apparent space group P21 with a unique axis (b) of 408.9 Å and with evidence of twinning, as indicated by the cumulative intensity distribution L statistic, suggesting the possibility of space group P1. Structure solution by molecular replacement shows a superhelical arrangement of six NONO homodimers (or 12 in P1) oriented parallel to the long axis, resulting in extensive noncrystallographic symmetry. Further analysis revealed that the crystal was not twinned, but the collected data suffered from highly overlapping reflections that obscured the L-test. Optimized data collection on a new crystal using higher energy X-rays, a smaller beam width and an increased sample-to-detector distance produced non-overlapping reflections to 2.6 Å resolution. The steps taken to analyse and overcome this series of practical difficulties and to produce a biologically informative structure are discussed.
Organizational Affiliation:
School of Chemistry and Biochemistry, The University of Western Australia, Crawley, WA 6009, Australia.