A High-Resolution Crystal Structure of a Psychrohalophilic alpha-Carbonic Anhydrase from Photobacterium profundum Reveals a Unique Dimer Interface.
Somalinga, V., Buhrman, G., Arun, A., Rose, R.B., Grunden, A.M.(2016) PLoS One 11: e0168022-e0168022
- PubMed: 27936100
- DOI: https://doi.org/10.1371/journal.pone.0168022
- Primary Citation of Related Structures:
5HPJ - PubMed Abstract:
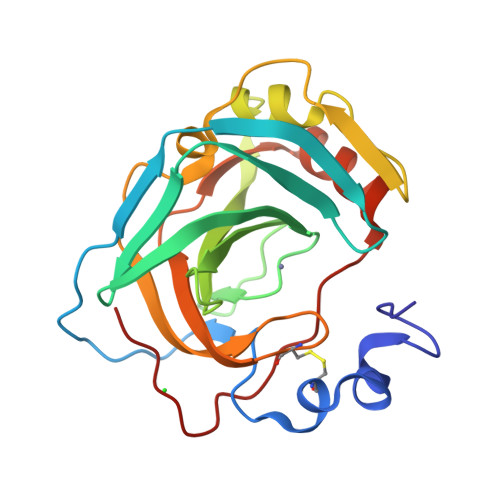
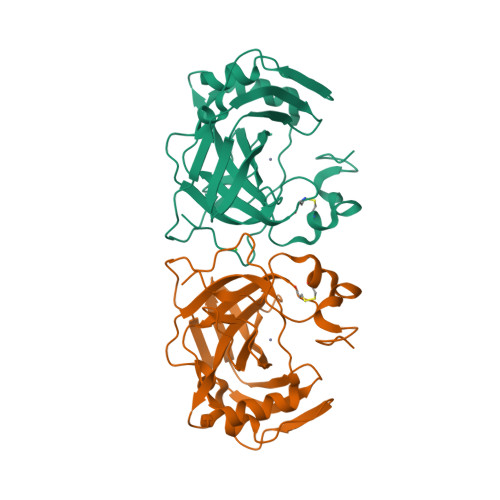
Bacterial α-carbonic anhydrases (α-CA) are zinc containing metalloenzymes that catalyze the rapid interconversion of CO2 to bicarbonate and a proton. We report the first crystal structure of a pyschrohalophilic α-CA from a deep-sea bacterium, Photobacterium profundum. Size exclusion chromatography of the purified P. profundum α-CA (PprCA) reveals that the protein is a heterogeneous mix of monomers and dimers. Furthermore, an "in-gel" carbonic anhydrase activity assay, also known as protonography, revealed two distinct bands corresponding to monomeric and dimeric forms of PprCA that are catalytically active. The crystal structure of PprCA was determined in its native form and reveals a highly conserved "knot-topology" that is characteristic of α-CA's. Similar to other bacterial α-CA's, PprCA also crystallized as a dimer. Furthermore, dimer interface analysis revealed the presence of a chloride ion (Cl-) in the interface which is unique to PprCA and has not been observed in any other α-CA's characterized so far. Molecular dynamics simulation and chloride ion occupancy analysis shows 100% occupancy for the Cl- ion in the dimer interface. Zinc coordinating triple histidine residues, substrate binding hydrophobic patch residues, and the hydrophilic proton wire residues are highly conserved in PprCA and are identical to other well-studied α-CA's.
Organizational Affiliation:
Department of Plant and Microbial Biology, North Carolina State University, Raleigh, NC, United States of America.