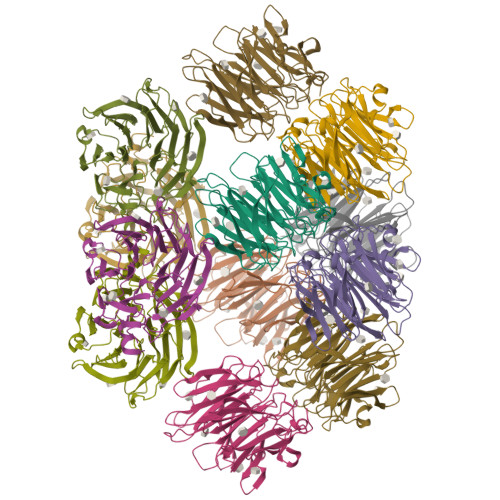
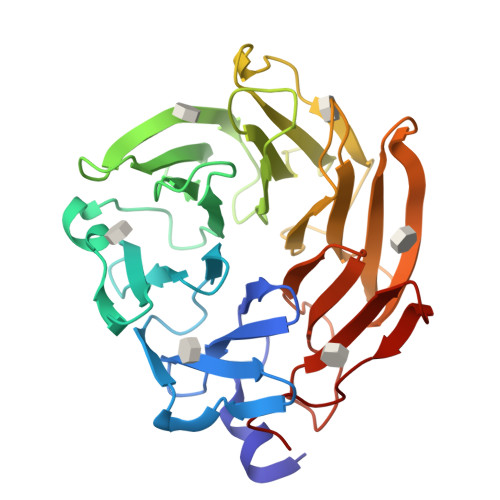
Synthesis of seleno-fucose compounds and their application to the X-ray structural determination of carbohydrate-lectin complexes using single/multi-wavelength anomalous dispersion phasing
Shimabukuro, J., Makyio, H., Suzuki, T., Nishikawa, Y., Kawasaki, M., Imamura, A., Ishida, H., Ando, H., Kato, R., Kiso, M.(2017) Bioorg Med Chem 25: 1132-1142
- PubMed: 28041800
- DOI: https://doi.org/10.1016/j.bmc.2016.12.021
- Primary Citation of Related Structures:
5H47 - PubMed Abstract:
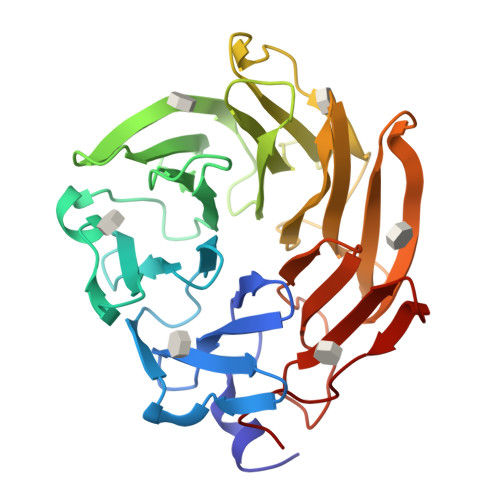
Selenium-incorporated fucoses (seleno-fucoses) differing in the position of the seleno-substituent were synthesized and applied to the X-ray structural determination of a carbohydrate-lectin complex using single/multi-wavelength anomalous dispersion (SAD/MAD) phasing. The hydroxyl groups at the C-1, -2, -3 and -4 position of fucose were individually substituted with a methylseleno group via a transacetalization reaction using MeSeCH 2 OBn or by an S N 2 reaction with TolSe - equivalents to afford the corresponding MeSe-fucose. The three-dimensional structures of a fucose-binding lectin complexed with several of these MeSe-fucoses have been determined by SAD/MAD phasing by utilizing the diffraction of selenium in the bound MeSe-fucoses.
Organizational Affiliation:
Department of Applied Bioorganic Chemistry, Gifu University, 1-1 Yanagido, Gifu 501-1193, Japan; Institute for Integrated Cell-Material Sciences (WPI-iCeMS), Kyoto University, Yoshida Ushinomiya-cho, Sakyo-ku, Kyoto 606-8501, Japan.