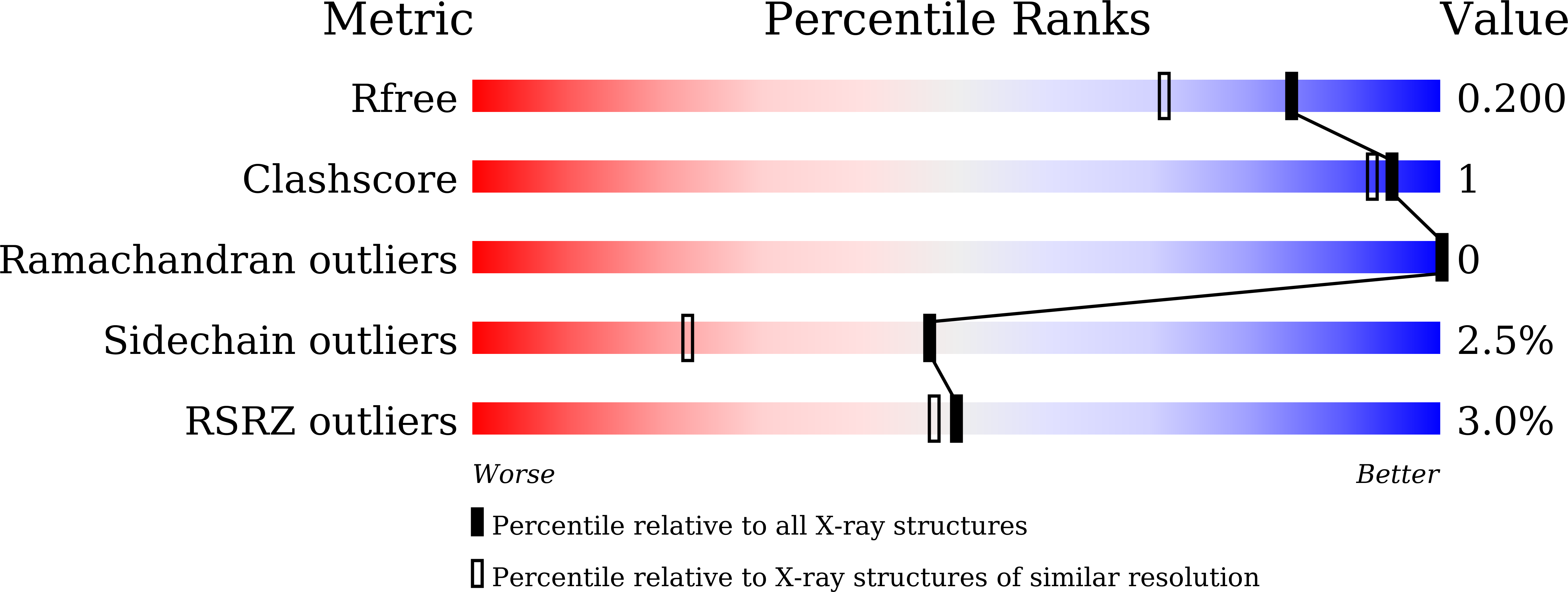
High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Yi, H., Choi, J.M., Hwang, J., Prati, F., Cao, T.P., Lee, S.H., Kim, H.S.(2016) Sci Rep 6: 36527-36527
- PubMed: 27827433
- DOI: https://doi.org/10.1038/srep36527
- Primary Citation of Related Structures:
5GL9, 5GLA, 5GLB, 5GLC, 5GLD - PubMed Abstract:
The omega loop in β-lactamases plays a pivotal role in substrate recognition and catalysis, and some mutations in this loop affect the adaptability of the enzymes to new antibiotics. Various mutations, including substitutions, deletions, and intragenic duplications resulting in tandem repeats (TRs), have been associated with β-lactamase substrate spectrum extension. TRs are unique among the mutations as they cause severe structural perturbations in the enzymes. We explored the process by which TRs are accommodated in order to test the adaptability of the omega loop. Structures of the mutant enzymes showed that the extra amino acid residues in the omega loop were freed outward from the enzyme, thereby maintaining the overall enzyme integrity. This structural adjustment was accompanied by disruptions of the internal α-helix and hydrogen bonds that originally maintained the conformation of the omega loop and the active site. Consequently, the mutant enzymes had a relaxed binding cavity, allowing for access of new substrates, which regrouped upon substrate binding in an induced-fit manner for subsequent hydrolytic reactions. Together, the data demonstrate that the design of the binding cavity, including the omega loop with its enormous adaptive capacity, is the foundation of the continuous evolution of β-lactamases against new drugs.
Organizational Affiliation:
Department of Biomedical Sciences, Korea University, Anam-Dong, Seongbuk-Gu, Seoul 136-705, Korea.