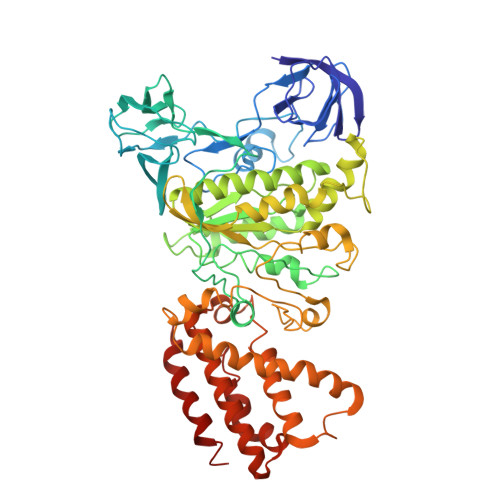
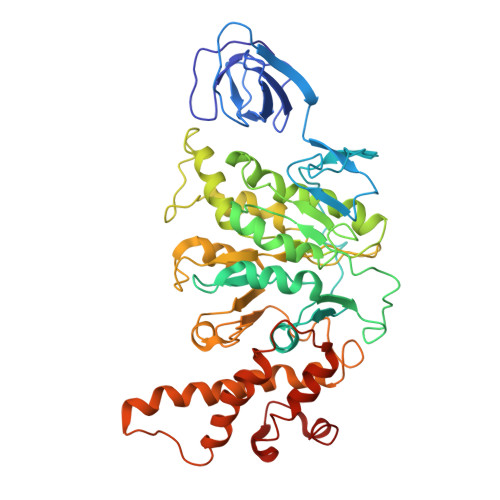
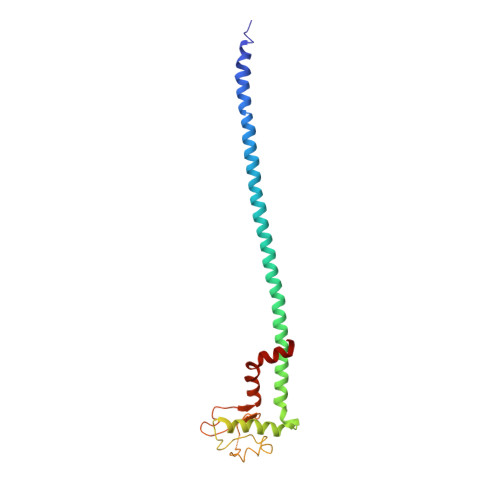
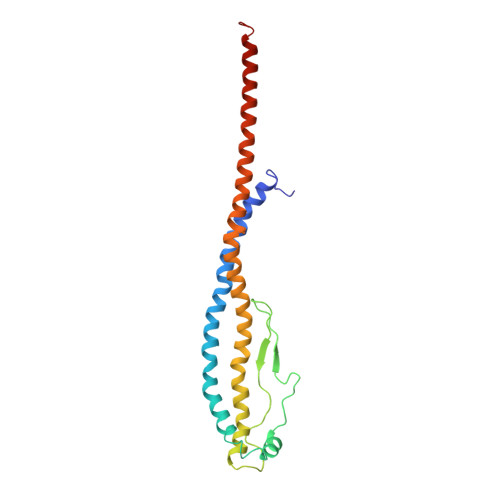
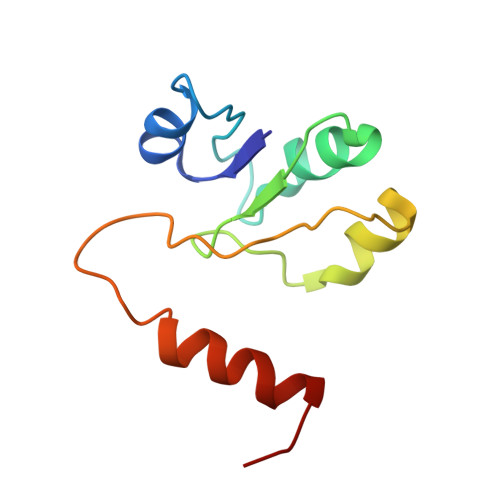
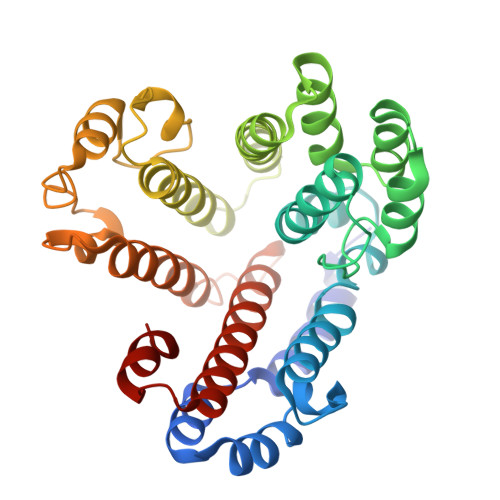
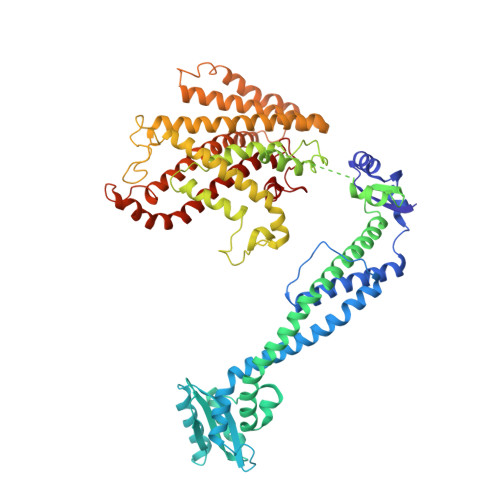
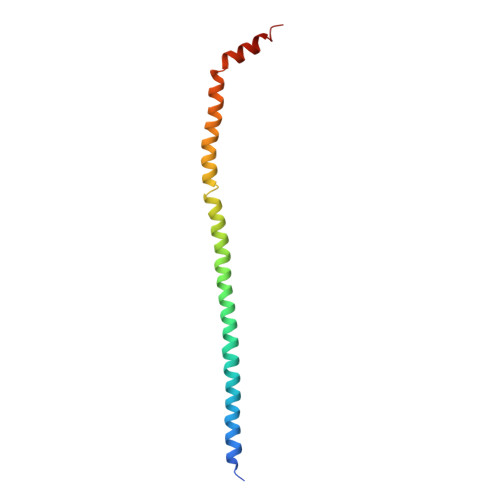Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Schep, D.G., Zhao, J., Rubinstein, J.L.(2016) Proc Natl Acad Sci U S A 113: 3245-3250
- PubMed: 26951669
- DOI: https://doi.org/10.1073/pnas.1521990113
- Primary Citation of Related Structures:
5GAR, 5GAS, 5I1M - PubMed Abstract:
Rotary ATPases couple ATP synthesis or hydrolysis to proton translocation across a membrane. However, understanding proton translocation has been hampered by a lack of structural information for the membrane-embedded a subunit. The V/A-ATPase from the eubacterium Thermus thermophilus is similar in structure to the eukaryotic V-ATPase but has a simpler subunit composition and functions in vivo to synthesize ATP rather than pump protons. We determined the T. thermophilus V/A-ATPase structure by cryo-EM at 6.4 Å resolution. Evolutionary covariance analysis allowed tracing of the a subunit sequence within the map, providing a complete model of the rotary ATPase. Comparing the membrane-embedded regions of the T. thermophilus V/A-ATPase and eukaryotic V-ATPase from Saccharomyces cerevisiae allowed identification of the α-helices that belong to the a subunit and revealed the existence of previously unknown subunits in the eukaryotic enzyme. Subsequent evolutionary covariance analysis enabled construction of a model of the a subunit in the S. cerevisae V-ATPase that explains numerous biochemical studies of that enzyme. Comparing the two a subunit structures determined here with a structure of the distantly related a subunit from the bovine F-type ATP synthase revealed a conserved pattern of residues, suggesting a common mechanism for proton transport in all rotary ATPases.
Organizational Affiliation:
The Hospital for Sick Children Research Institute, Toronto, ON, Canada M5G0A4; Department of Medical Biophysics, The University of Toronto, Toronto, ON, Canada M5G1L7;