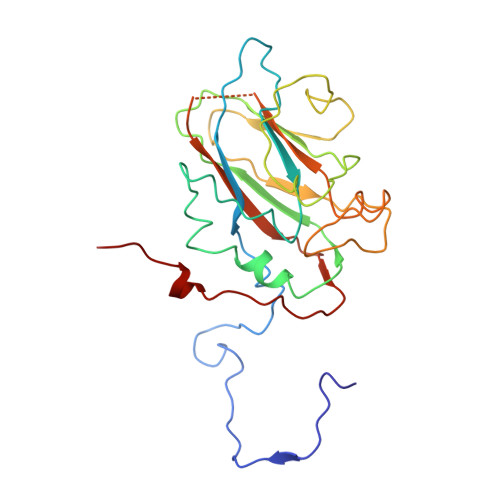
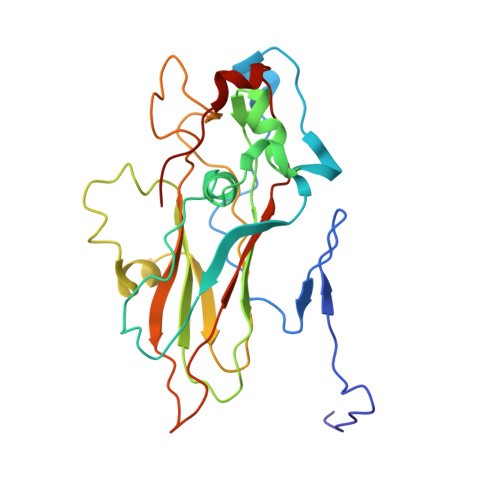
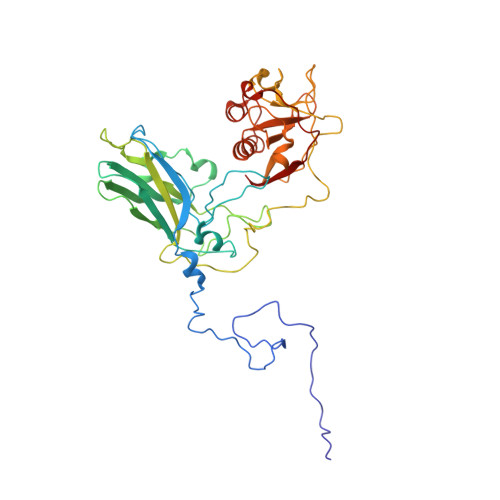
Structure of deformed wing virus, a major honey bee pathogen.
Skubnik, K., Novacek, J., Fuzik, T., Pridal, A., Paxton, R.J., Plevka, P.(2017) Proc Natl Acad Sci U S A 114: 3210-3215
- PubMed: 28270616
- DOI: https://doi.org/10.1073/pnas.1615695114
- Primary Citation of Related Structures:
5G51, 5G52, 5L7Q, 5L8Q, 5MUP, 5MV5, 5MV6 - PubMed Abstract:
The worldwide population of western honey bees ( Apis mellifera ) is under pressure from habitat loss, environmental stress, and pathogens, particularly viruses that cause lethal epidemics. Deformed wing virus (DWV) from the family Iflaviridae , together with its vector, the mite Varroa destructor , is likely the major threat to the world's honey bees. However, lack of knowledge of the atomic structures of iflaviruses has hindered the development of effective treatments against them. Here, we present the virion structures of DWV determined to a resolution of 3.1 Å using cryo-electron microscopy and 3.8 Å by X-ray crystallography. The C-terminal extension of capsid protein VP3 folds into a globular protruding (P) domain, exposed on the virion surface. The P domain contains an Asp-His-Ser catalytic triad that is, together with five residues that are spatially close, conserved among iflaviruses. These residues may participate in receptor binding or provide the protease, lipase, or esterase activity required for entry of the virus into a host cell. Furthermore, nucleotides of the DWV RNA genome interact with VP3 subunits. The capsid protein residues involved in the RNA binding are conserved among honey bee iflaviruses, suggesting a putative role of the genome in stabilizing the virion or facilitating capsid assembly. Identifying the RNA-binding and putative catalytic sites within the DWV virion structure enables future analyses of how DWV and other iflaviruses infect insect cells and also opens up possibilities for the development of antiviral treatments.
Organizational Affiliation:
Structural Virology, Central European Institute of Technology, Masaryk University, 62500 Brno, Czech Republic.