Structure and Organization of Heteromeric Ampa-Type Glutamate Receptors.
Herguedas, B., Garcia-Nafria, J., Cais, O., Fernandez-Leiro, R., Krieger, J., Ho, H., Greger, I.H.(2016) Science 352: 3873
- PubMed: 26966189
- DOI: https://doi.org/10.1126/science.aad3873
- Primary Citation of Related Structures:
5FWX, 5FWY, 5IDE, 5IDF - PubMed Abstract:
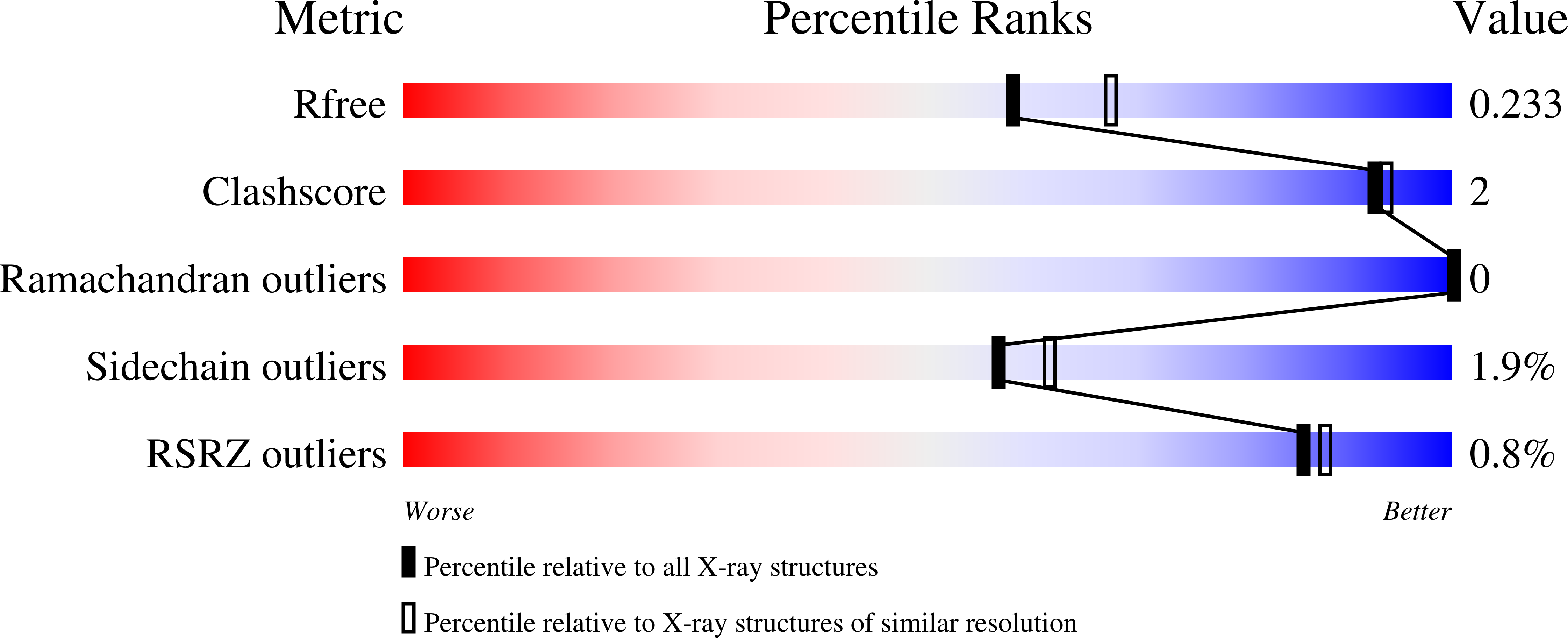
AMPA-type glutamate receptors (AMPARs), which are central mediators of rapid neurotransmission and synaptic plasticity, predominantly exist as heteromers of the subunits GluA1 to GluA4. Here we report the first AMPAR heteromer structures, which deviate substantially from existing GluA2 homomer structures. Crystal structures of the GluA2/3 and GluA2/4 N-terminal domains reveal a novel compact conformation with an alternating arrangement of the four subunits around a central axis. This organization is confirmed by cysteine cross-linking in full-length receptors, and it permitted us to determine the structure of an intact GluA2/3 receptor by cryogenic electron microscopy. Two models in the ligand-free state, at resolutions of 8.25 and 10.3 angstroms, exhibit substantial vertical compression and close associations between domain layers, reminiscent of N-methyl-D-aspartate receptors. Model 1 resembles a resting state and model 2 a desensitized state, thus providing snapshots of gating transitions in the nominal absence of ligand. Our data reveal organizational features of heteromeric AMPARs and provide a framework to decipher AMPAR architecture and signaling.
Organizational Affiliation:
Neurobiology Division, MRC Laboratory of Molecular Biology, Cambridge, UK.