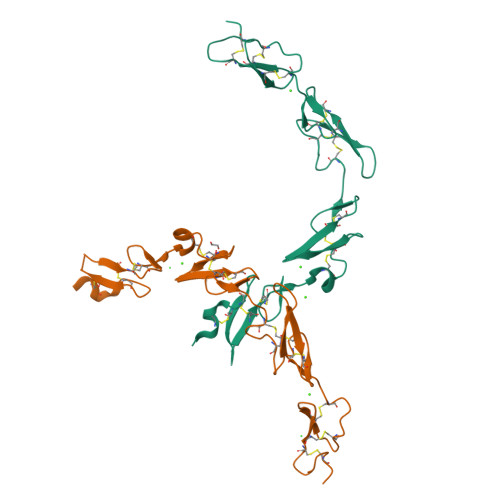
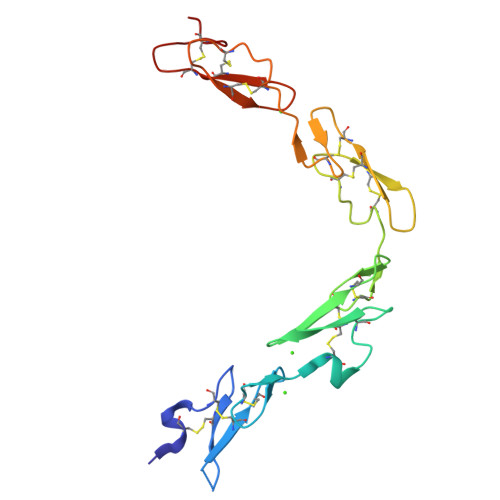
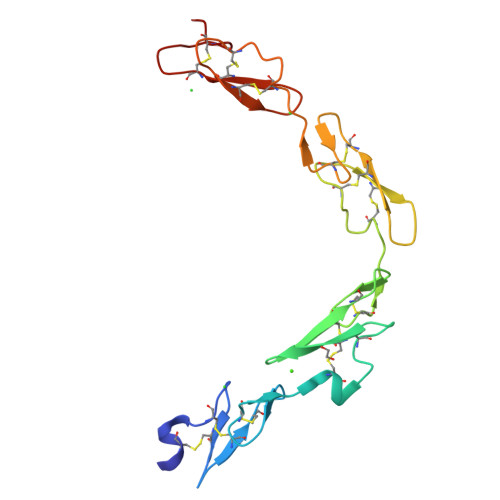
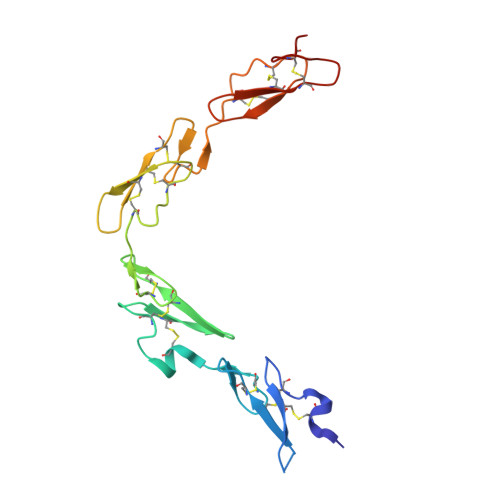
Non-Linear and Flexible Regions of the Human Notch1 Extracellular Domain Revealed by High-Resolution Structural Studies.
Weisshuhn, P.C., Sheppard, D., Taylor, P., Whiteman, P., Lea, S.M., Handford, P.A., Redfield, C.(2016) Structure 24: 555
- PubMed: 26996961
- DOI: https://doi.org/10.1016/j.str.2016.02.010
- Primary Citation of Related Structures:
5FM9, 5FMA - PubMed Abstract:
The Notch receptor is a key component of a core metazoan signaling pathway activated by Delta/Serrate/Lag-2 ligands expressed on an adjacent cell. This results in a short-range signal with profound effects on cell-fate determination, cell proliferation, and cell death. Key to understanding receptor function is structural knowledge of the large extracellular portion of Notch which contains multiple repeats of epidermal growth factor (EGF)-like domains. Here we investigate the EGF4-13 region of human Notch1 (hN1) using a multidisciplinary approach. Ca(2+)-binding measurements, X-ray crystallography, {(1)H}-(15)N heteronuclear nuclear Overhauser effects, and residual dipolar couplings support a non-linear organization for the EGF4-13 region with a rigid, bent conformation for EGF4-7 and a single flexible linkage between EGF9 and EGF10. These data allow us to construct an informed model for EGF10-13 which, in conjunction with comparative binding studies, demonstrates that EGF10 has an important role in determining Notch receptor sensitivity to Dll-4.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.