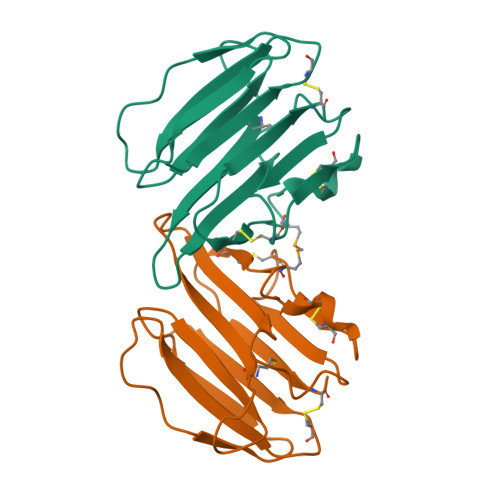
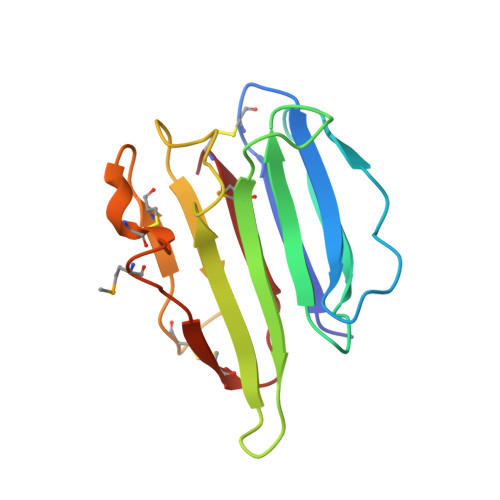
Crystal Structure Analysis and the Identification of Distinctive Functional Regions of the Protein Elicitor Mohrip2
Liu, M., Duan, L., Wang, M., Zeng, H., Liu, X., Qiu, D.(2016) Front Plant Sci 7: 1103-1103
- PubMed: 27507984
- DOI: https://doi.org/10.3389/fpls.2016.01103
- Primary Citation of Related Structures:
5FID - PubMed Abstract:
The protein elicitor MoHrip2, which was extracted from Magnaporthe oryzae as an exocrine protein, triggers the tobacco immune system and enhances blast resistance in rice. However, the detailed mechanisms by which MoHrip2 acts as an elicitor remain unclear. Here, we investigated the structure of MoHrip2 to elucidate its functions based on molecular structure. The three-dimensional structure of MoHrip2 was obtained. Overall, the crystal structure formed a β-barrel structure and showed high similarity to the pathogenesis-related (PR) thaumatin superfamily protein thaumatin-like xylanase inhibitor (TL-XI). To investigate the functional regions responsible for MoHrip2 elicitor activities, the full length and eight truncated proteins were expressed in Escherichia coli and were evaluated for elicitor activity in tobacco. Biological function analysis showed that MoHrip2 triggered the defense system against Botrytis cinerea in tobacco. Moreover, only MoHrip2M14 and other fragments containing the 14 amino acids residues in the middle region of the protein showed the elicitor activity of inducing a hypersensitive response and resistance related pathways, which were similar to that of full-length MoHrip2. These results revealed that the central 14 amino acid residues were essential for anti-pathogenic activity.
Organizational Affiliation:
State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection - Chinese Academy of Agricultural Sciences Beijing, China.