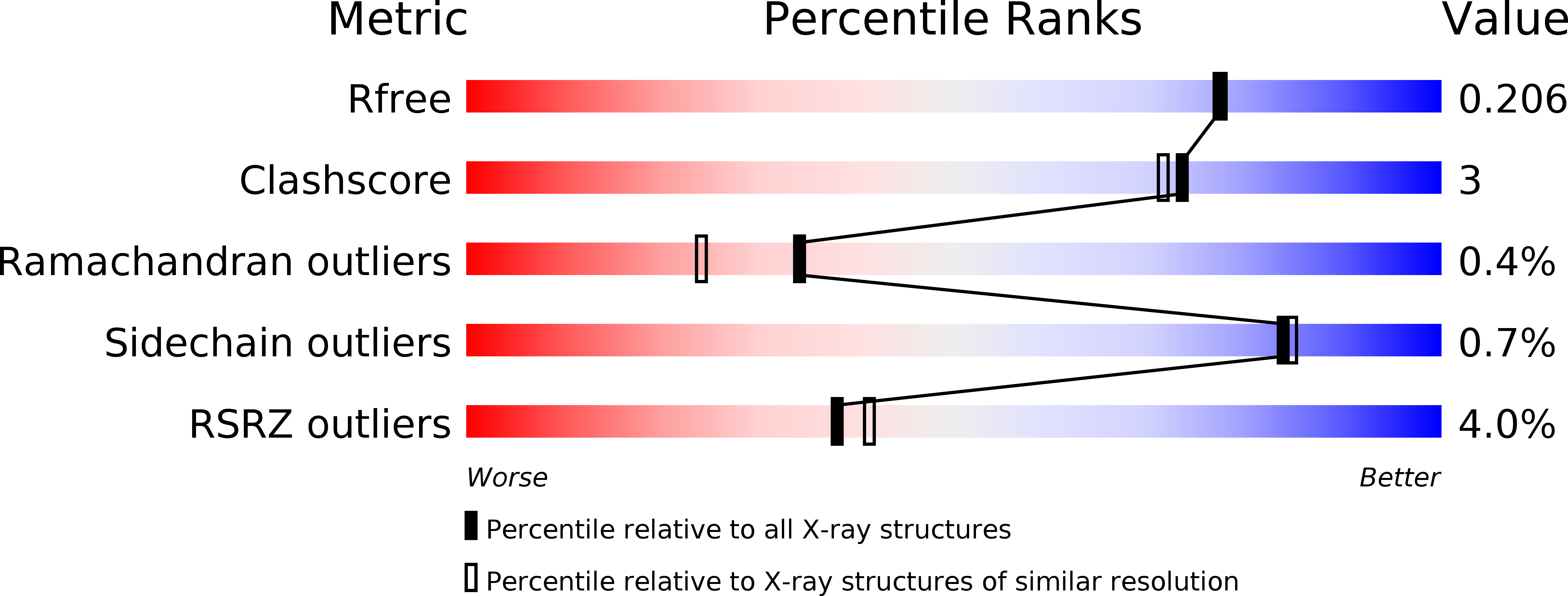
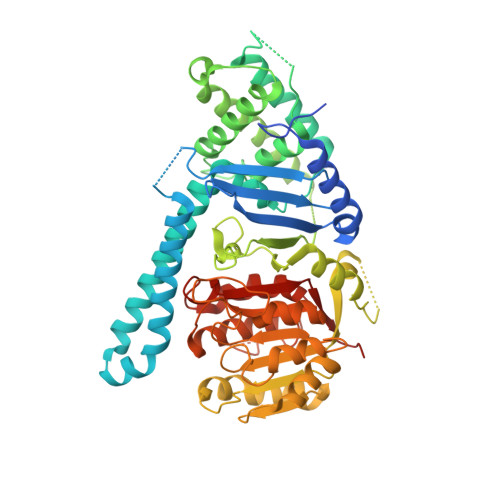
Structural basis of cohesin cleavage by separase.
Lin, Z., Luo, X., Yu, H.(2016) Nature 532: 131-134
- PubMed: 27027290
- DOI: https://doi.org/10.1038/nature17402
- Primary Citation of Related Structures:
5FBY, 5FC2, 5FC3 - PubMed Abstract:
Accurate chromosome segregation requires timely dissolution of chromosome cohesion after chromosomes are properly attached to the mitotic spindle. Separase is absolutely essential for cohesion dissolution in organisms from yeast to man. It cleaves the kleisin subunit of cohesin and opens the cohesin ring to allow chromosome segregation. Cohesin cleavage is spatiotemporally controlled by separase-associated regulatory proteins, including the inhibitory chaperone securin, and by phosphorylation of both the enzyme and substrates. Dysregulation of this process causes chromosome missegregation and aneuploidy, contributing to cancer and birth defects. Despite its essential functions, atomic structures of separase have not been determined. Here we report crystal structures of the separase protease domain from the thermophilic fungus Chaetomium thermophilum, alone or covalently bound to unphosphorylated and phosphorylated inhibitory peptides derived from a cohesin cleavage site. These structures reveal how separase recognizes cohesin and how cohesin phosphorylation by polo-like kinase 1 (Plk1) enhances cleavage. Consistent with a previous cellular study, mutating two securin residues in a conserved motif that partly matches the separase cleavage consensus converts securin from a separase inhibitor to a substrate. Our study establishes atomic mechanisms of substrate cleavage by separase and suggests competitive inhibition by securin.
Organizational Affiliation:
Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, 6001 Forest Park Road, Dallas, Texas 75390, USA.