Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex
Li, J., He, Y., Lu, Q., Zhang, M.(2016) Dev Cell 36: 179-189
- PubMed: 26812017
- DOI: https://doi.org/10.1016/j.devcel.2015.12.020
- Primary Citation of Related Structures:
5F3X, 5F3Y - PubMed Abstract:
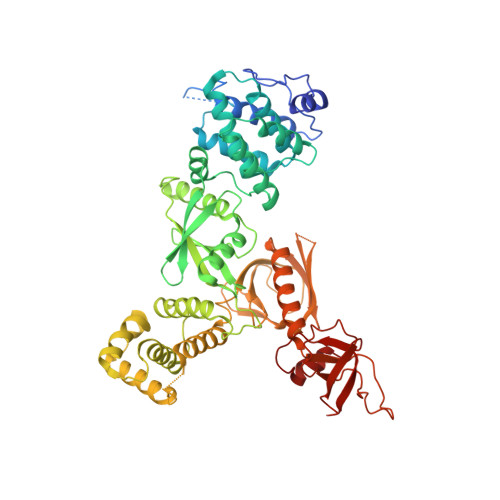
Brush border microvilli are actin-based protrusions lining the apical surface of epithelial cells in intestines and proximal tubules of kidneys. While brush border microvilli resemble the relatively well-characterized stereocilia of hair cells, the mechanistic basis of tip-link complex organization in microvilli is poorly understood. Here, we have biochemically and structurally characterized the following pairs of interactions: protocadherin 24 and Harmonin (also known as USH1C or AIE-75), Harmonin and myosin VIIb (MYO7B), Harmonin and ANKS4B, and ANKS4B and MYO7B. We show that Harmonin, ANKS4B, and MYO7B form a stable ternary complex for anchoring microvilli tip-link cadherins. Despite having only Harmonin in common, the microvilli and the stereocilia tip-link complexes are formed via strikingly similar interaction modes. These results not only provide insight into the mechanistic bases of brush border microvilli formation and maintenance but may also be valuable for understanding some gut and/or kidney diseases caused by perturbations of brush border microvilli structures.
Organizational Affiliation:
Division of Life Science, State Key Laboratory of Molecular Neuroscience, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, China.