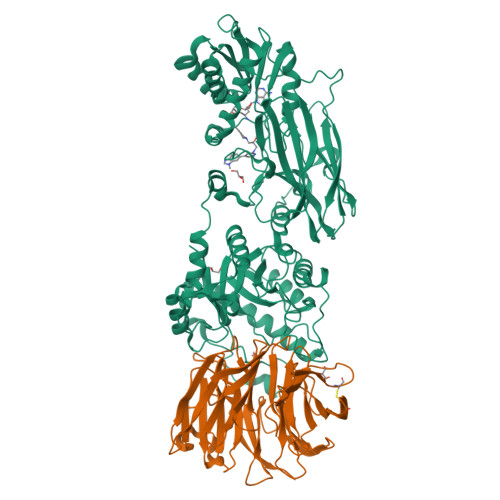
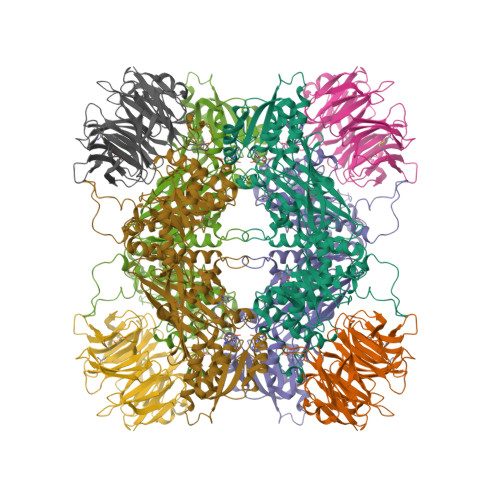
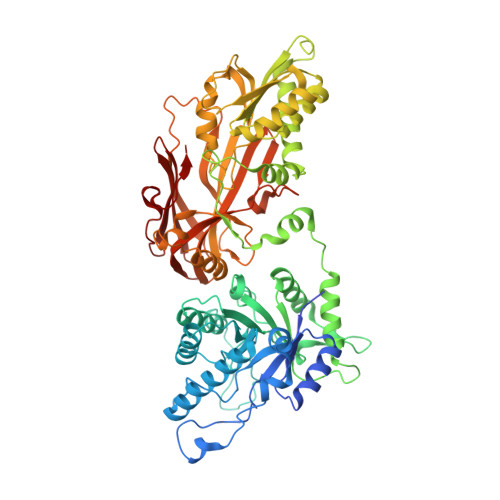
Structure and Property Guided Design in the Identification of PRMT5 Tool Compound EPZ015666.
Duncan, K.W., Rioux, N., Boriack-Sjodin, P.A., Munchhof, M.J., Reiter, L.A., Majer, C.R., Jin, L., Johnston, L.D., Chan-Penebre, E., Kuplast, K.G., Porter Scott, M., Pollock, R.M., Waters, N.J., Smith, J.J., Moyer, M.P., Copeland, R.A., Chesworth, R.(2016) ACS Med Chem Lett 7: 162-166
- PubMed: 26985292
- DOI: https://doi.org/10.1021/acsmedchemlett.5b00380
- Primary Citation of Related Structures:
5EMJ, 5EMK, 5EML, 5EMM - PubMed Abstract:
The recent publication of a potent and selective inhibitor of protein methyltransferase 5 (PRMT5) provides the scientific community with in vivo-active tool compound EPZ015666 (GSK3235025) to probe the underlying pharmacology of this key enzyme. Herein, we report the design and optimization strategies employed on an initial hit compound with poor in vitro clearance to yield in vivo tool compound EPZ015666 and an additional potent in vitro tool molecule EPZ015866 (GSK3203591).
Organizational Affiliation:
Epizyme, Inc. , 400 Technology Square, Cambridge, Massachusetts 02139, United States.