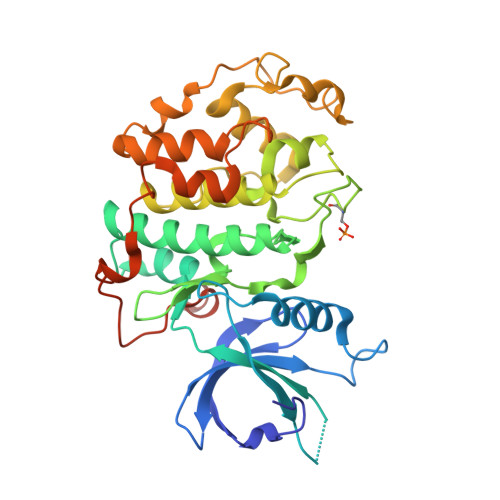
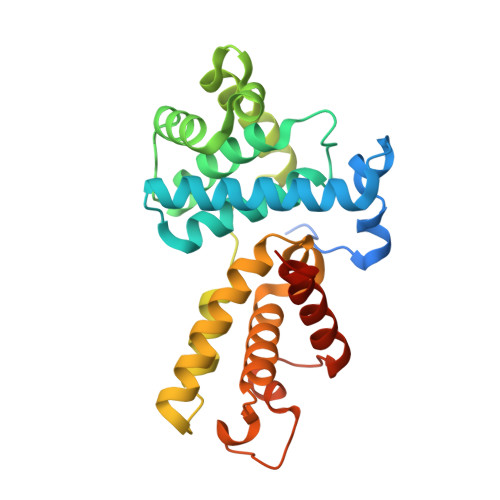
Structural and Functional Analysis of the Cdk13/Cyclin K Complex.
Greifenberg, A.K., Honig, D., Pilarova, K., Duster, R., Bartholomeeusen, K., Bosken, C.A., Anand, K., Blazek, D., Geyer, M.(2016) Cell Rep 14: 320-331
- PubMed: 26748711
- DOI: https://doi.org/10.1016/j.celrep.2015.12.025
- Primary Citation of Related Structures:
5EFQ - PubMed Abstract:
Cyclin-dependent kinases regulate the cell cycle and transcription in higher eukaryotes. We have determined the crystal structure of the transcription kinase Cdk13 and its Cyclin K subunit at 2.0 Å resolution. Cdk13 contains a C-terminal extension helix composed of a polybasic cluster and a DCHEL motif that interacts with the bound ATP. Cdk13/CycK phosphorylates both Ser5 and Ser2 of the RNA polymerase II C-terminal domain (CTD) with a preference for Ser7 pre-phosphorylations at a C-terminal position. The peptidyl-prolyl isomerase Pin1 does not change the phosphorylation specificities of Cdk9, Cdk12, and Cdk13 but interacts with the phosphorylated CTD through its WW domain. Using recombinant proteins, we find that flavopiridol inhibits Cdk7 more potently than it does Cdk13. Gene expression changes after knockdown of Cdk13 or Cdk12 are markedly different, with enrichment of growth signaling pathways for Cdk13-dependent genes. Together, our results provide insights into the structure, function, and activity of human Cdk13/CycK.
Organizational Affiliation:
Institute of Innate Immunity, Department of Structural Immunology, University of Bonn, Sigmund-Freud-Strasse 25, 53127 Bonn, Germany; Center of Advanced European Studies and Research, Group Physical Biochemistry, Ludwig-Erhard-Allee 2, 53175 Bonn, Germany.