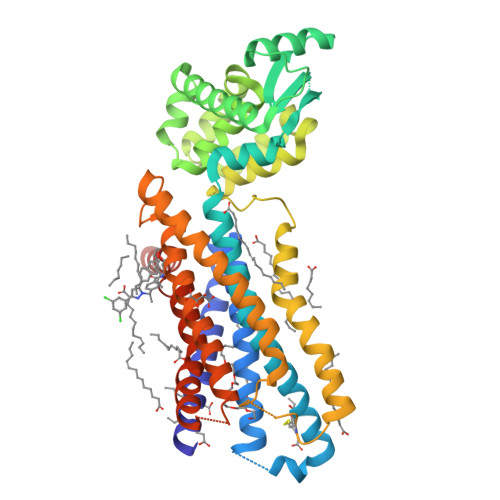
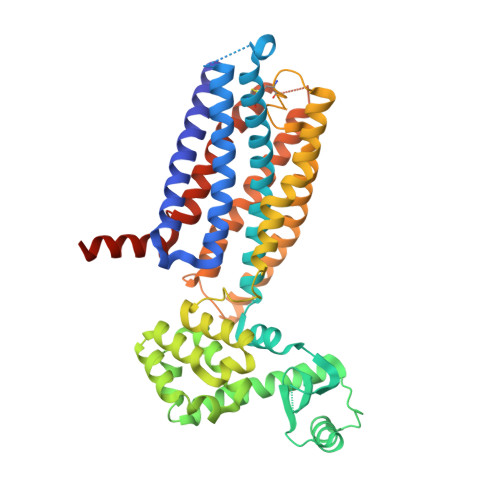
Extra-helical binding site of a glucagon receptor antagonist.
Jazayeri, A., Dore, A.S., Lamb, D., Krishnamurthy, H., Southall, S.M., Baig, A.H., Bortolato, A., Koglin, M., Robertson, N.J., Errey, J.C., Andrews, S.P., Teobald, I., Brown, A.J., Cooke, R.M., Weir, M., Marshall, F.H.(2016) Nature 533: 274-277
- PubMed: 27111510
- DOI: https://doi.org/10.1038/nature17414
- Primary Citation of Related Structures:
5EE7 - PubMed Abstract:
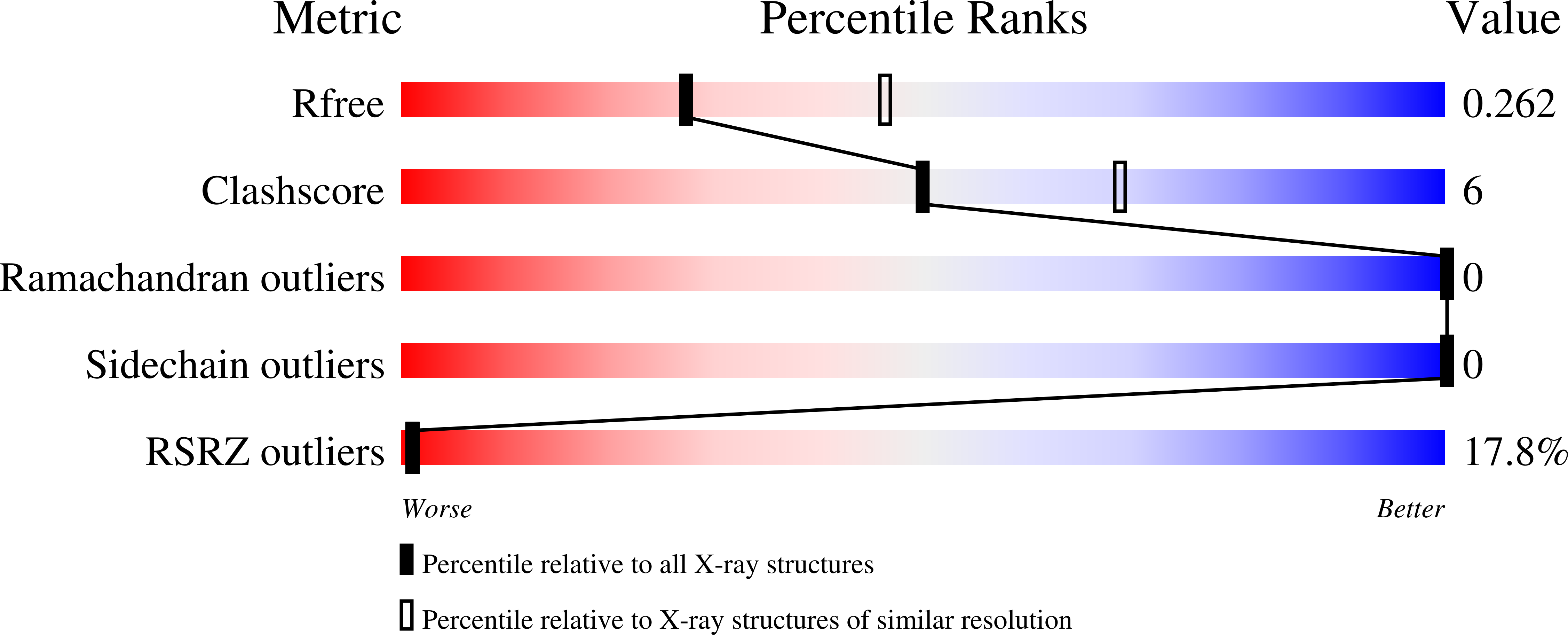
Glucagon is a 29-amino-acid peptide released from the α-cells of the islet of Langerhans, which has a key role in glucose homeostasis. Glucagon action is transduced by the class B G-protein-coupled glucagon receptor (GCGR), which is located on liver, kidney, intestinal smooth muscle, brain, adipose tissue, heart and pancreas cells, and this receptor has been considered an important drug target in the treatment of diabetes. Administration of recently identified small-molecule GCGR antagonists in patients with type 2 diabetes results in a substantial reduction of fasting and postprandial glucose concentrations. Although an X-ray structure of the transmembrane domain of the GCGR has previously been solved, the ligand (NNC0640) was not resolved. Here we report the 2.5 Å structure of human GCGR in complex with the antagonist MK-0893 (ref. 4), which is found to bind to an allosteric site outside the seven transmembrane (7TM) helical bundle in a position between TM6 and TM7 extending into the lipid bilayer. Mutagenesis of key residues identified in the X-ray structure confirms their role in the binding of MK-0893 to the receptor. The unexpected position of the binding site for MK-0893, which is structurally similar to other GCGR antagonists, suggests that glucagon activation of the receptor is prevented by restriction of the outward helical movement of TM6 required for G-protein coupling. Structural knowledge of class B receptors is limited, with only one other ligand-binding site defined--for the corticotropin-releasing hormone receptor 1 (CRF1R)--which was located deep within the 7TM bundle. We describe a completely novel allosteric binding site for class B receptors, providing an opportunity for structure-based drug design for this receptor class and furthering our understanding of the mechanisms of activation of these receptors.
Organizational Affiliation:
Heptares Therapeutics Ltd, BioPark, Broadwater Road, Welwyn Garden City, Hertfordshire AL7 3AX, UK.