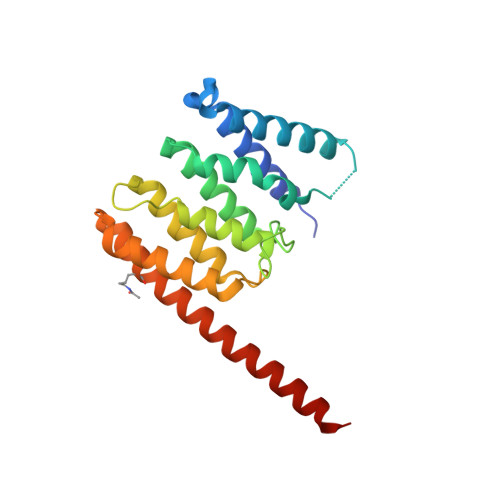
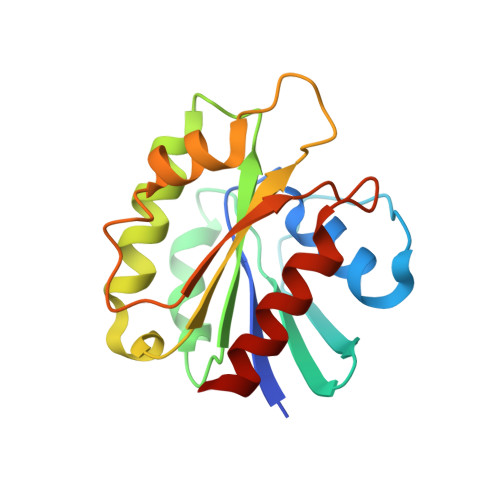
Structural Insights into Arl1-Mediated Targeting of the Arf-GEF BIG1 to the trans-Golgi.
Galindo, A., Soler, N., McLaughlin, S.H., Yu, M., Williams, R.L., Munro, S.(2016) Cell Rep 16: 839-850
- PubMed: 27373159
- DOI: https://doi.org/10.1016/j.celrep.2016.06.022
- Primary Citation of Related Structures:
5EE5 - PubMed Abstract:
The GTPase Arf1 is the major regulator of vesicle traffic at both the cis- and trans-Golgi. Arf1 is activated at the cis-Golgi by the guanine nucleotide exchange factor (GEF) GBF1 and at the trans-Golgi by the related GEF BIG1 or its paralog, BIG2. The trans-Golgi-specific targeting of BIG1 and BIG2 depends on the Arf-like GTPase Arl1. We find that Arl1 binds to the dimerization and cyclophilin binding (DCB) domain in BIG1 and report a crystal structure of human Arl1 bound to this domain. Residues in the DCB domain that bind Arl1 are required for BIG1 to locate to the Golgi in vivo. DCB domain-binding residues in Arl1 have a distinct conformation from those in known Arl1-effector complexes, and this plasticity allows Arl1 to interact with different effectors of unrelated structure. The findings provide structural insight into how Arf1 GEFs, and hence active Arf1, achieve their correct subcellular distribution.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge CB2 0QH, UK.