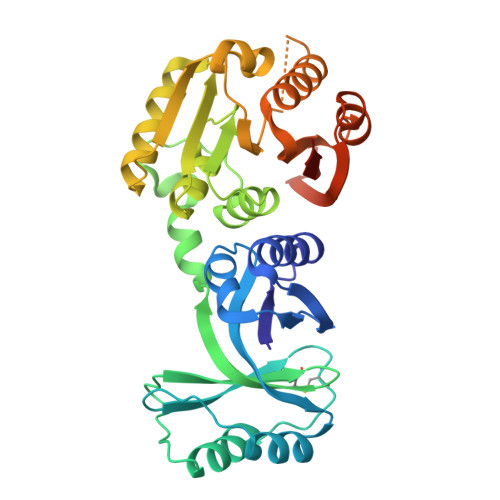
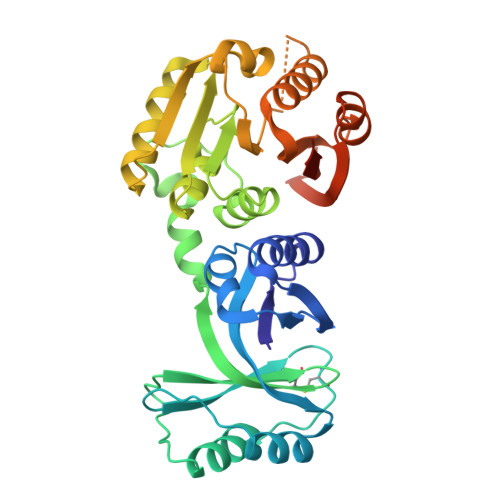
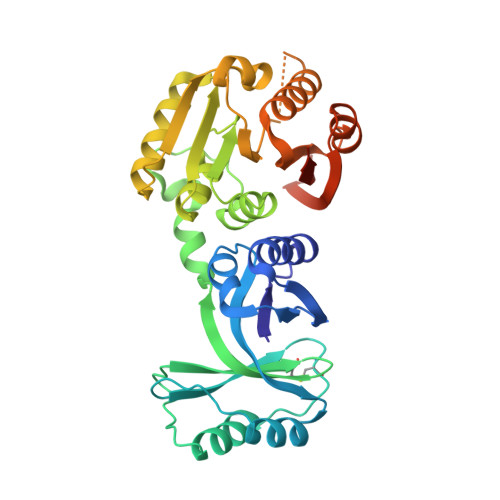
Structural and functional analyses of the archaeal tRNA m2G/m22G10 methyltransferase aTrm11 provide mechanistic insights into site specificity of a tRNA methyltransferase that contains common RNA-binding modules.
Hirata, A., Nishiyama, S., Tamura, T., Yamauchi, A., Hori, H.(2016) Nucleic Acids Res
- PubMed: 27325738
- DOI: https://doi.org/10.1093/nar/gkw561
- Primary Citation of Related Structures:
5E71, 5E72 - PubMed Abstract:
N(2)-methylguanosine is one of the most universal modified nucleosides required for proper function in transfer RNA (tRNA) molecules. In archaeal tRNA species, a specific S-adenosyl-L-methionine (SAM)-dependent tRNA methyltransferase (MTase), aTrm11, catalyzes formation of N(2)-methylguanosine and N(2),N(2)-dimethylguanosine at position 10. Here, we report the first X-ray crystal structures of aTrm11 from Thermococcus kodakarensis (Tko), of the apo-form, and of its complex with SAM. The structures show that TkoTrm11 consists of three domains: an N-terminal ferredoxinlike domain (NFLD), THUMP domain and Rossmann-fold MTase (RFM) domain. A linker region connects the THUMP-NFLD and RFM domains. One SAM molecule is bound in the pocket of the RFM domain, suggesting that TkoTrm11 uses a catalytic mechanism similar to that of other tRNA MTases containing an RFM domain. Furthermore, the conformation of NFLD and THUMP domains in TkoTrm11 resembles that of other tRNA-modifying enzymes specifically recognizing the tRNA acceptor stem. Our docking model of TkoTrm11-SAM in complex with tRNA, combined with biochemical analyses and pre-existing evidence, provides insights into the substrate tRNA recognition mechanism: The THUMP domain recognizes a 3'-ACCA end, and the linker region and RFM domain recognize the T-stem, acceptor stem and V-loop of tRNA, thereby causing TkoTrm11 to specifically identify its methylation site.
Organizational Affiliation:
Department of Materials Science and Biotechnology, Graduate School of Science and Engineering, Ehime University, 3 Bunkyo-cho, Matsuyama, Ehime 790-8577, Japan ahirata@ehime-u.ac.jp.