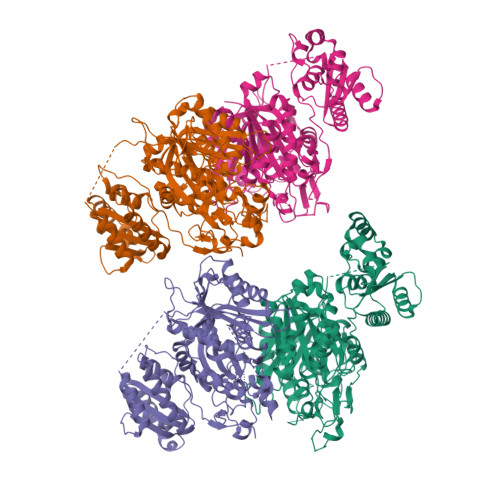
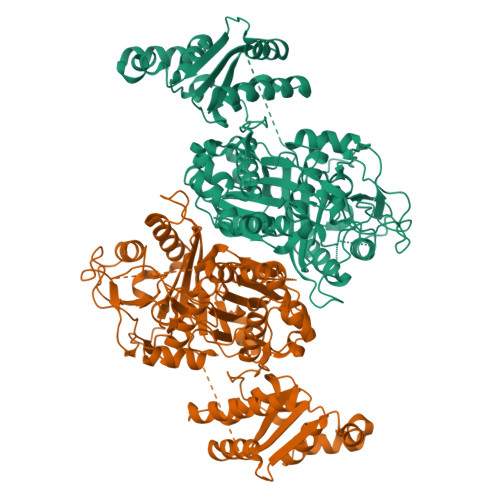
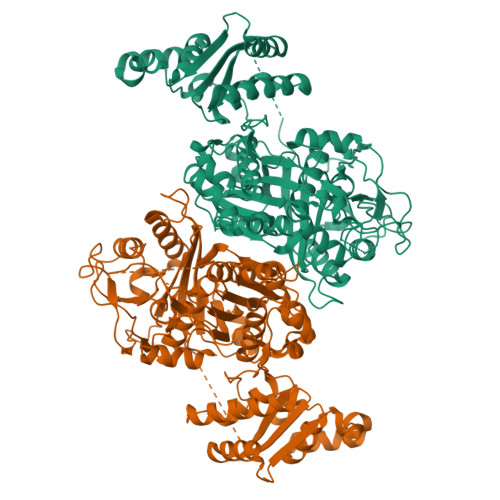
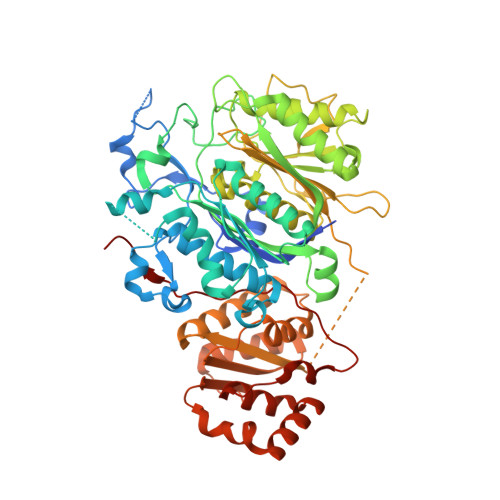
The LINKS motif zippers trans-acyltransferase polyketide synthase assembly lines into a biosynthetic megacomplex.
Gay, D.C., Wagner, D.T., Meinke, J.L., Zogzas, C.E., Gay, G.R., Keatinge-Clay, A.T.(2016) J Struct Biol 193: 196-205
- PubMed: 26724270
- DOI: https://doi.org/10.1016/j.jsb.2015.12.011
- Primary Citation of Related Structures:
5E5N, 5E6K, 5ELP, 5ENY, 5ERB, 5ERF - PubMed Abstract:
Polyketides such as the clinically-valuable antibacterial agent mupirocin are constructed by architecturally-sophisticated assembly lines known as trans-acyltransferase polyketide synthases. Organelle-sized megacomplexes composed of several copies of trans-acyltransferase polyketide synthase assembly lines have been observed by others through transmission electron microscopy to be located at the Bacillus subtilis plasma membrane, where the synthesis and export of the antibacterial polyketide bacillaene takes place. In this work we analyze ten crystal structures of trans-acyltransferase polyketide synthases ketosynthase domains, seven of which are reported here for the first time, to characterize a motif capable of zippering assembly lines into a megacomplex. While each of the three-helix LINKS (Laterally-INteracting Ketosynthase Sequence) motifs is observed to similarly dock with a spatially-reversed copy of itself through hydrophobic and ionic interactions, the amino acid sequences of this motif are not conserved. Such a code is appropriate for mediating homotypic contacts between assembly lines to ensure the ordered self-assembly of a noncovalent, yet tightly-knit, enzymatic network. LINKS-mediated lateral interactions would also have the effect of bolstering the vertical association of the polypeptides that comprise a polyketide synthase assembly line.
Organizational Affiliation:
Department of Molecular Biosciences, The University of Texas at Austin, 1 University Station A5000, Austin, TX 78712-0165, United States.