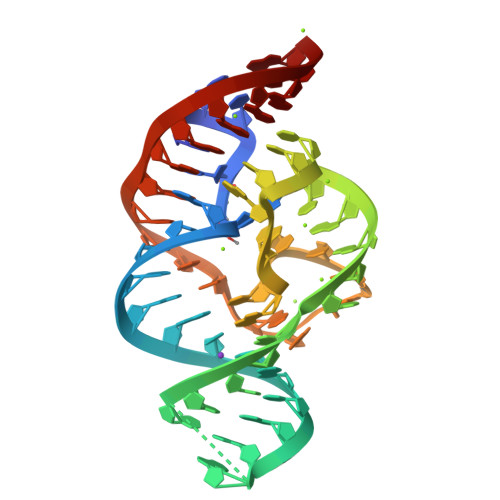
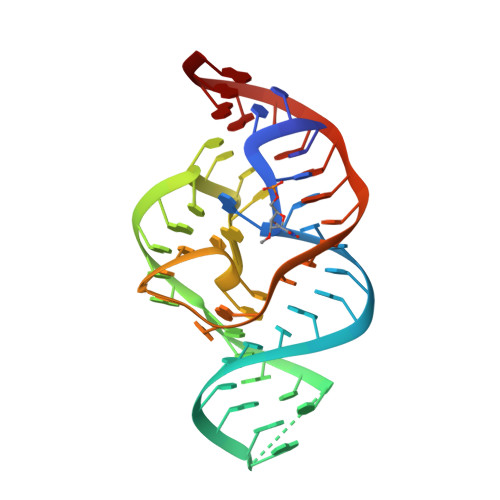
A Mini-Twister Variant and Impact of Residues/Cations on the Phosphodiester Cleavage of this Ribozyme Class.
Kosutic, M., Neuner, S., Ren, A., Flur, S., Wunderlich, C., Mairhofer, E., Vusurovic, N., Seikowski, J., Breuker, K., Hobartner, C., Patel, D.J., Kreutz, C., Micura, R.(2015) Angew Chem Int Ed Engl 54: 15128-15133
- PubMed: 26473980
- DOI: https://doi.org/10.1002/anie.201506601
- Primary Citation of Related Structures:
5DUN - PubMed Abstract:
Nucleolytic ribozymes catalyze site-specific cleavage of their phosphodiester backbones. A minimal version of the twister ribozyme is reported that lacks the phylogenetically conserved stem P1 while retaining wild-type activity. Atomic mutagenesis revealed that nitrogen atoms N1 and N3 of the adenine-6 at the cleavage site are indispensable for cleavage. By NMR spectroscopy, a pKa value of 5.1 was determined for a (13) C2-labeled adenine at this position in the twister ribozyme, which is significantly shifted compared to the pKa of the same adenine in the substrate alone. This finding pinpoints at a potential role for adenine-6 in the catalytic mechanism besides the previously identified invariant guanine-48 and a Mg(2+) ion, both of which are directly coordinated to the non-bridging oxygen atoms of the scissile phosphate; for the latter, additional evidence stems from the observation that Mn(2+) or Cd(2+) accelerated cleavage of phosphorothioate substrates. The relevance of this metal ion binding site is further emphasized by a new 2.6 Å X-ray structure of a 2'-OCH3 -U5 modified twister ribozyme.
Organizational Affiliation:
Institute of Organic Chemistry and Center for Molecular Biosciences CMBI, Leopold-Franzens University, Innrain 80-82, 6020 Innsbruck (Austria).