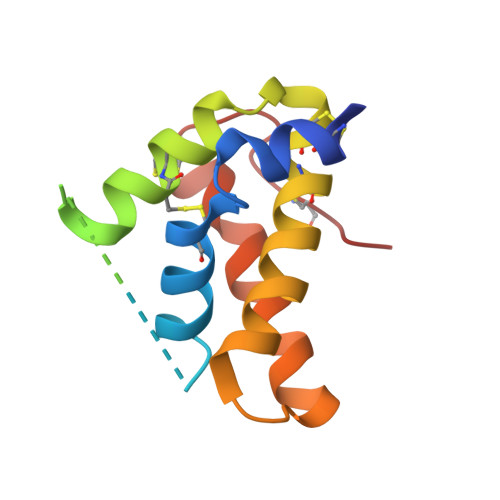
Crystal structure of mature 2S albumin from Moringa oleifera seeds.
Ullah, A., Mariutti, R.B., Masood, R., Caruso, I.P., Gravatim Costa, G.H., Millena de Freita, C., Santos, C.R., Zanphorlin, L.M., Rossini Mutton, M.J., Murakami, M.T., Arni, R.K.(2015) Biochem Biophys Res Commun 468: 365-371
- PubMed: 26505799
- DOI: https://doi.org/10.1016/j.bbrc.2015.10.087
- Primary Citation of Related Structures:
5DOM - PubMed Abstract:
2S albumins, the seed storage proteins, are the primary sources of carbon and nitrogen and are involved in plant defense. The mature form of Moringa oleifera (M. oleifera), a chitin binding protein isoform 3-1 (mMo-CBP3-1) a thermostable antifungal, antibacterial, flocculating 2S albumin is widely used for the treatment of water and is potentially interesting for the development of both antifungal drugs and transgenic crops. The crystal structure of mMo-CBP3-1 determined at 1.7 Å resolution demonstrated that it is comprised of two proteolytically processed α-helical chains, stabilized by four disulfide bridges that is stable, resistant to pH changes and has a melting temperature (TM) of approximately 98 °C. The surface arginines and the polyglutamine motif are the key structural factors for the observed flocculating, antibacterial and antifungal activities. This represents the first crystal structure of a 2S albumin and the model of the pro-protein indicates the structural changes that occur upon formation of mMo-CBP3-1 and determines the structural motif and charge distribution patterns for the diverse observed activities.
- Multiuser Center for Biomolecular Innovation, Department of Physics, IBILCE/UNESP, São Jose do Rio Preto, SP, Brazil. Electronic address: anwar.ms90@yahoo.com.
Organizational Affiliation: