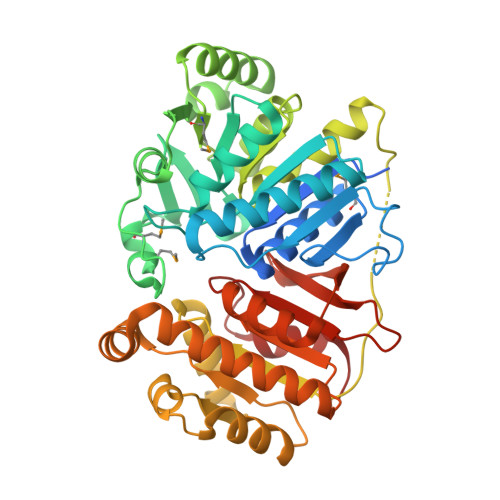
Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP.
Vetting, M.W., Al Obaidi, N.F., Morisco, L.L., Benach, J., Wasserman, S.R., Attonito, J.D., Chamala, S., Chowdhury, S., Love, J., Seidel, R.D., Whalen, K.L., Gerlt, J.A., Almo, S.C., Enzyme Function Initiative (EFI)To be published.