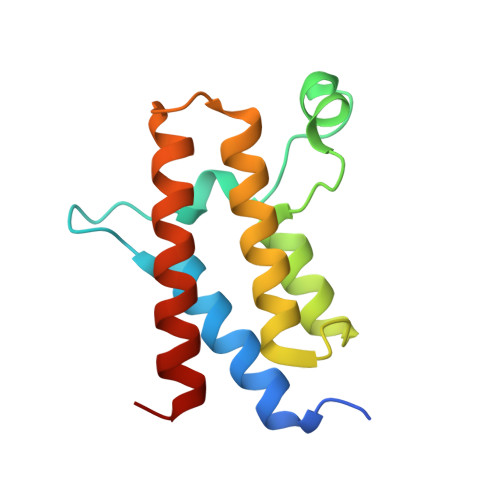
Crystal structure of the bromodomain of human BRM (SMARCA2) in complex with a hydroxyphenyl propenone inhibitor 17
Tallant, C., Owen, D.R., Taylor, A., Fedorov, O., Savitsky, P., Siejka, P., Srikannathasan, V., Nowak, R., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Knapp, S.To be published.