Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Zhang, J., Yang, Q., Romero, J.A., Cross, J., Wang, B., Poutsiaka, K.M., Epie, F., Bevan, D., Wu, Y., Moy, T., Daniel, A., Chamberlain, B., Carter, N., Shotwell, J., Arya, A., Kumar, V., Silverman, J., Nguyen, K., Metcalf, C.A., Ryan, D., Lippa, B., Dolle, R.E.(2015) ACS Med Chem Lett 6: 1080-1085
- PubMed: 26487916
- DOI: https://doi.org/10.1021/acsmedchemlett.5b00266
- Primary Citation of Related Structures:
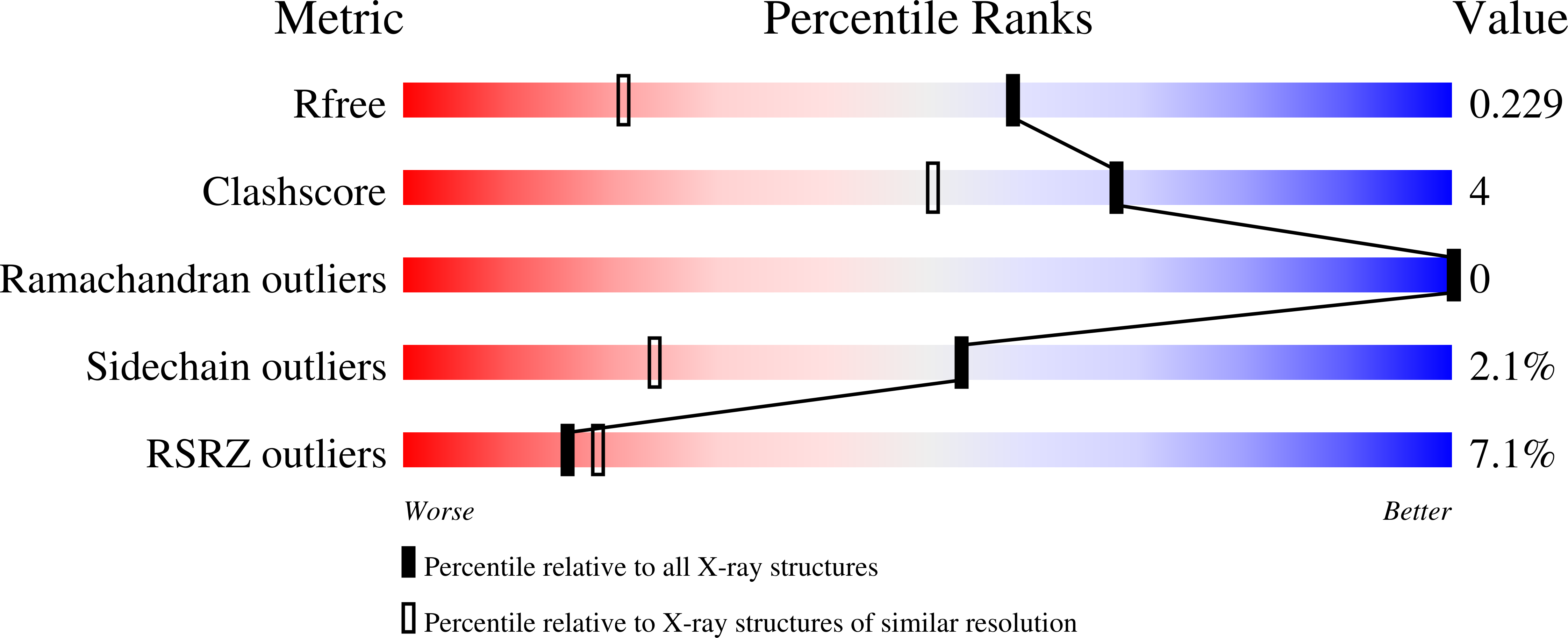
5D7D, 5D7R - PubMed Abstract:
Antibacterials with a novel mechanism of action offer a great opportunity to combat widespread antimicrobial resistance. Bacterial DNA Gyrase is a clinically validated target. Through physiochemical property optimization of a pyrazolopyridone hit, a novel class of GyrB inhibitors were discovered. Guided by structure-based drug design, indazole derivatives with excellent enzymatic and antibacterial activity as well as great animal efficacy were discovered.
Organizational Affiliation:
Cubist Pharmaceuticals Inc. , Lexington, Massachusetts 02421, United States.