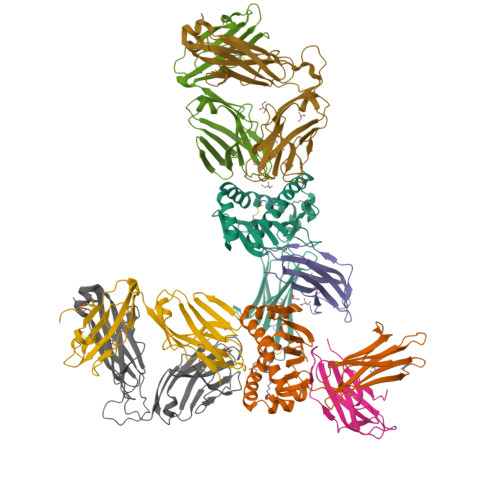
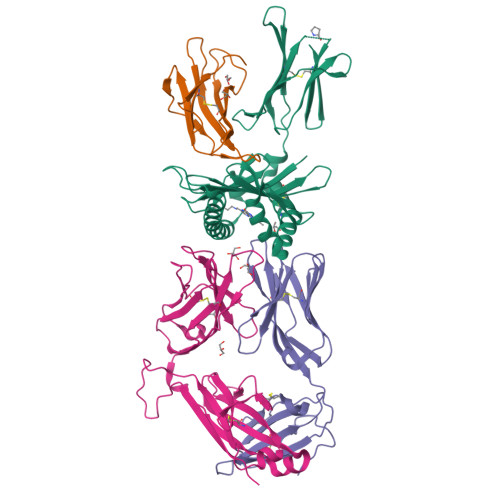
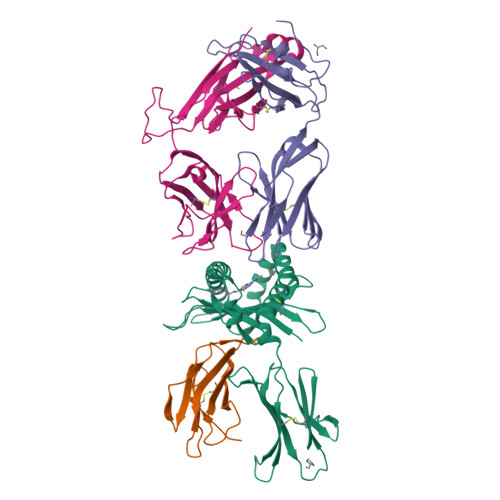
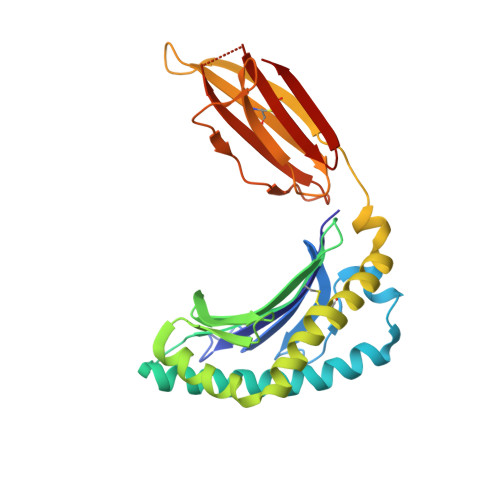
Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Gherardin, N.A., Keller, A.N., Woolley, R.E., Le Nours, J., Ritchie, D.S., Neeson, P.J., Birkinshaw, R.W., Eckle, S.B., Waddington, J.N., Liu, L., Fairlie, D.P., Uldrich, A.P., Pellicci, D.G., McCluskey, J., Godfrey, D.I., Rossjohn, J.(2016) Immunity 44: 32-45
- PubMed: 26795251
- DOI: https://doi.org/10.1016/j.immuni.2015.12.005
- Primary Citation of Related Structures:
5D5M, 5D7I, 5D7J, 5D7K, 5D7L - PubMed Abstract:
A characteristic of mucosal-associated invariant T (MAIT) cells is the expression of TRAV1-2(+) T cell receptors (TCRs) that are activated by riboflavin metabolite-based antigens (Ag) presented by the MHC-I related molecule, MR1. Whether the MR1-restricted T cell repertoire and associated Ag responsiveness extends beyond these cells remains unclear. Here, we describe MR1 autoreactivity and folate-derivative reactivity in a discrete subset of TRAV1-2(+) MAIT cells. This recognition was attributable to CDR3β loop-mediated effects within a consensus TRAV1-2(+) TCR-MR1-Ag footprint. Furthermore, we have demonstrated differential folate- and riboflavin-derivative reactivity by a diverse population of "atypical" TRAV1-2(-) MR1-restricted T cells. We have shown that TRAV1-2(-) T cells are phenotypically heterogeneous and largely distinct from TRAV1-2(+) MAIT cells. A TRAV1-2(-) TCR docks more centrally on MR1, thereby adopting a markedly different molecular footprint to the TRAV1-2(+) TCR. Accordingly, diversity within the MR1-restricted T cell repertoire leads to differing MR1-restricted Ag specificity.
Organizational Affiliation:
Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, University of Melbourne, Parkville, Victoria 3010, Australia; Cancer Immunology Program, Peter MacCallum Cancer Centre, East Melbourne, Victoria 3002, Australia; ARC Centre of Excellence in Advanced Molecular Imaging, University of Melbourne, Parkville, Victoria 3010, Australia.