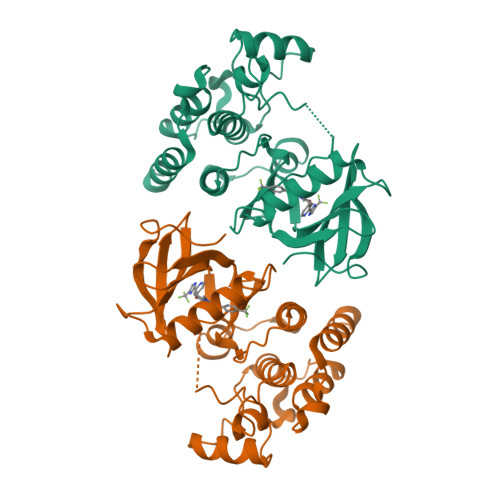
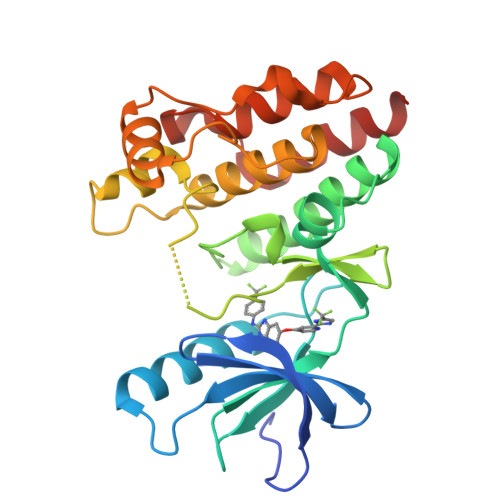
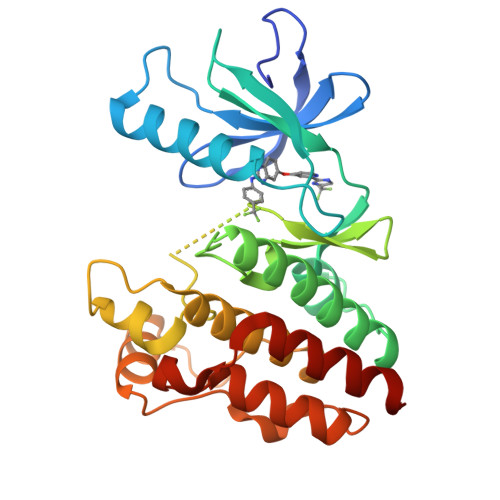
Discovery of RAF265: A Potent mut-B-RAF Inhibitor for the Treatment of Metastatic Melanoma.
Williams, T.E., Subramanian, S., Verhagen, J., McBride, C.M., Costales, A., Sung, L., Antonios-McCrea, W., McKenna, M., Louie, A.K., Ramurthy, S., Levine, B., Shafer, C.M., Machajewski, T., Renhowe, P.A., Appleton, B.A., Amiri, P., Chou, J., Stuart, D., Aardalen, K., Poon, D.(2015) ACS Med Chem Lett 6: 961-965
- PubMed: 26396681
- DOI: https://doi.org/10.1021/ml500526p
- Primary Citation of Related Structures:
5CT7 - PubMed Abstract:
Abrogation of errant signaling along the MAPK pathway through the inhibition of B-RAF kinase is a validated approach for the treatment of pathway-dependent cancers. We report the development of imidazo-benzimidazoles as potent B-RAF inhibitors. Robust in vivo efficacy coupled with correlating pharmacokinetic/pharmacodynamic (PKPD) and PD-efficacy relationships led to the identification of RAF265, 1, which has advanced into clinical trials.
Organizational Affiliation:
Global Discovery Chemistry, Oncology and Exploratory Chemistry, Novartis Institutes for Biomedical Research , 5300 Chiron Way, Emeryville, California 94608, United States.