Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with F9 ligand and the product L-Tryptophan in the beta-site.
Hilario, E., Caulkins, B.G., Young, R.P., Dunn, M.F., Mueller, L.J., Fan, L.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tryptophan synthase alpha chain | 268 | Salmonella enterica subsp. enterica serovar Typhimurium | Mutation(s): 0 Gene Names: trpA EC: 4.2.1.20 | 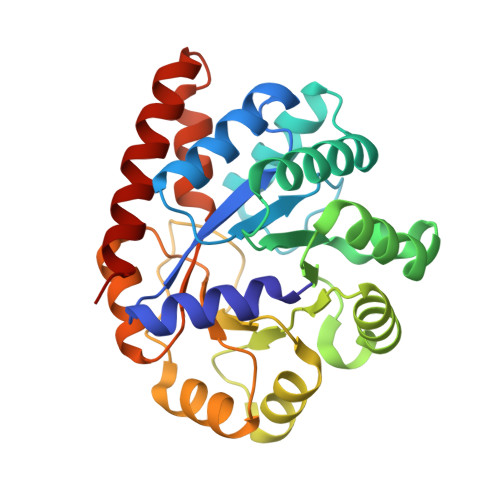 | |
UniProt | |||||
Find proteins for P00929 (Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)) Explore P00929 Go to UniProtKB: P00929 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00929 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tryptophan synthase beta chain | 397 | Salmonella enterica subsp. enterica serovar Typhimurium | Mutation(s): 0 Gene Names: trpB EC: 4.2.1.20 | 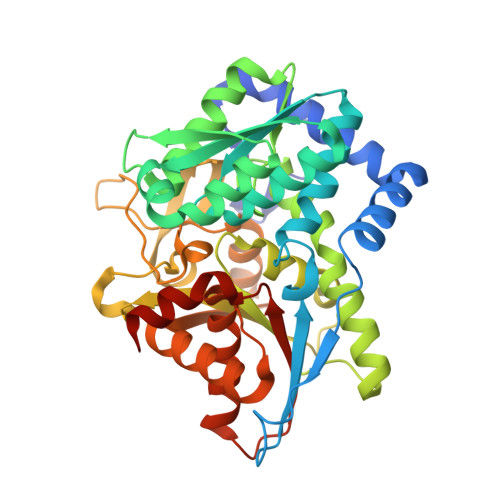 | |
UniProt | |||||
Find proteins for P0A2K1 (Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)) Explore P0A2K1 Go to UniProtKB: P0A2K1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A2K1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| F9F Query on F9F | C [auth A] | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE C9 H11 F3 N O7 P S JDDKDMFCTOZVCJ-UHFFFAOYSA-N |  | ||
| PLP Query on PLP | E [auth B] | PYRIDOXAL-5'-PHOSPHATE C8 H10 N O6 P NGVDGCNFYWLIFO-UHFFFAOYSA-N |  | ||
| TRP Query on TRP | F [auth B] | TRYPTOPHAN C11 H12 N2 O2 QIVBCDIJIAJPQS-VIFPVBQESA-N |  | ||
| BCN Query on BCN | G [auth B] | BICINE C6 H13 N O4 FSVCELGFZIQNCK-UHFFFAOYSA-N |  | ||
| CS Query on CS | D [auth A], H [auth B], I [auth B] | CESIUM ION Cs NCMHKCKGHRPLCM-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | J [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 182.553 | α = 90 |
| b = 59.3 | β = 94.82 |
| c = 67.368 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |
| MOLREP | phasing |
| DM | phasing |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01GM097569 |