Structural basis for gene regulation by a B12-dependent photoreceptor.
Jost, M., Fernandez-Zapata, J., Polanco, M.C., Ortiz-Guerrero, J.M., Chen, P.Y., Kang, G., Padmanabhan, S., Elias-Arnanz, M., Drennan, C.L.(2015) Nature 526: 536-541
- PubMed: 26416754
- DOI: https://doi.org/10.1038/nature14950
- Primary Citation of Related Structures:
5C8A, 5C8D, 5C8E, 5C8F - PubMed Abstract:
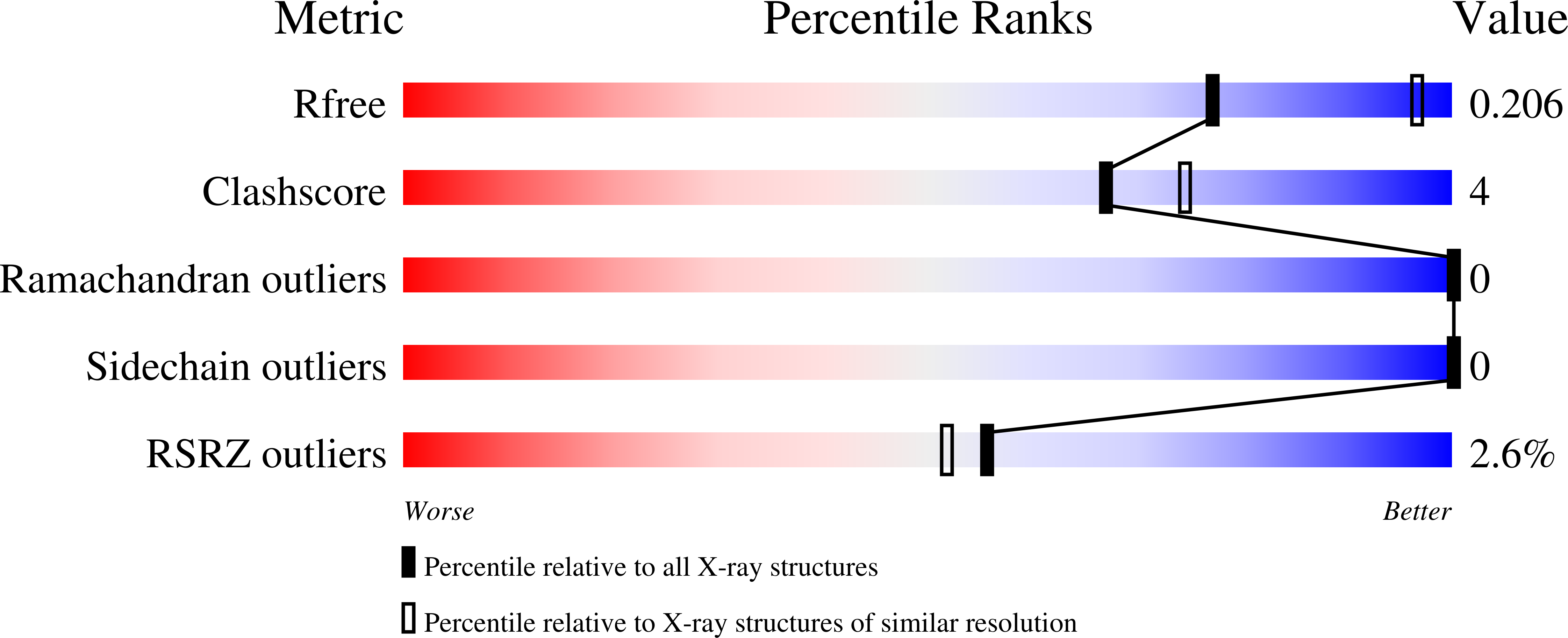
Photoreceptor proteins enable organisms to sense and respond to light. The newly discovered CarH-type photoreceptors use a vitamin B12 derivative, adenosylcobalamin, as the light-sensing chromophore to mediate light-dependent gene regulation. Here we present crystal structures of Thermus thermophilus CarH in all three relevant states: in the dark, both free and bound to operator DNA, and after light exposure. These structures provide visualizations of how adenosylcobalamin mediates CarH tetramer formation in the dark, how this tetramer binds to the promoter -35 element to repress transcription, and how light exposure leads to a large-scale conformational change that activates transcription. In addition to the remarkable functional repurposing of adenosylcobalamin from an enzyme cofactor to a light sensor, we find that nature also repurposed two independent protein modules in assembling CarH. These results expand the biological role of vitamin B12 and provide fundamental insight into a new mode of light-dependent gene regulation.
Organizational Affiliation:
Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, Massachusetts 02139, USA.