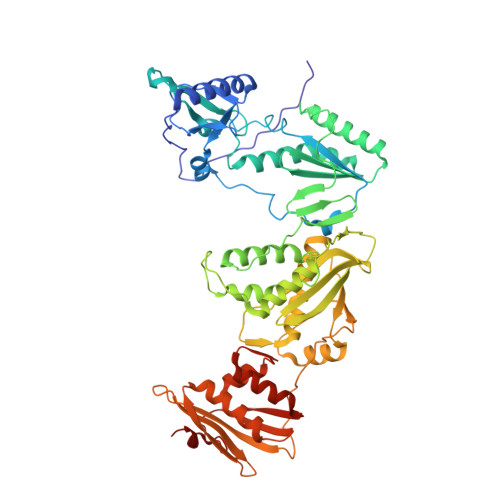
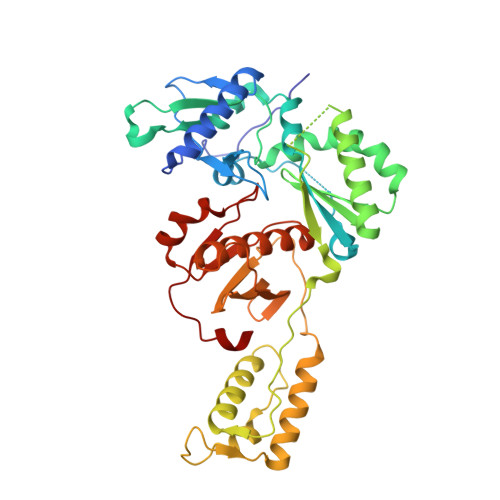
Discovery and crystallography of bicyclic arylaminoazines as potent inhibitors of HIV-1 reverse transcriptase.
Lee, W.G., Frey, K.M., Gallardo-Macias, R., Spasov, K.A., Chan, A.H., Anderson, K.S., Jorgensen, W.L.(2015) Bioorg Med Chem Lett 25: 4824-4827
- PubMed: 26166629
- DOI: https://doi.org/10.1016/j.bmcl.2015.06.074
- Primary Citation of Related Structures:
5C24, 5C25 - PubMed Abstract:
Non-nucleoside inhibitors of HIV-1 reverse transcriptase (HIV-RT) are reported that incorporate a 7-indolizinylamino or 2-naphthylamino substituent on a pyrimidine or 1,3,5-triazine core. The most potent compounds show below 10 nanomolar activity towards wild-type HIV-1 and variants bearing Tyr181Cys and Lys103Asn/Tyr181Cys resistance mutations. The compounds also feature good aqueous solubility. Crystal structures for two complexes enhance the analysis of the structure-activity data.
Organizational Affiliation:
Department of Chemistry, Yale University, New Haven, CT 06520-8107, USA.