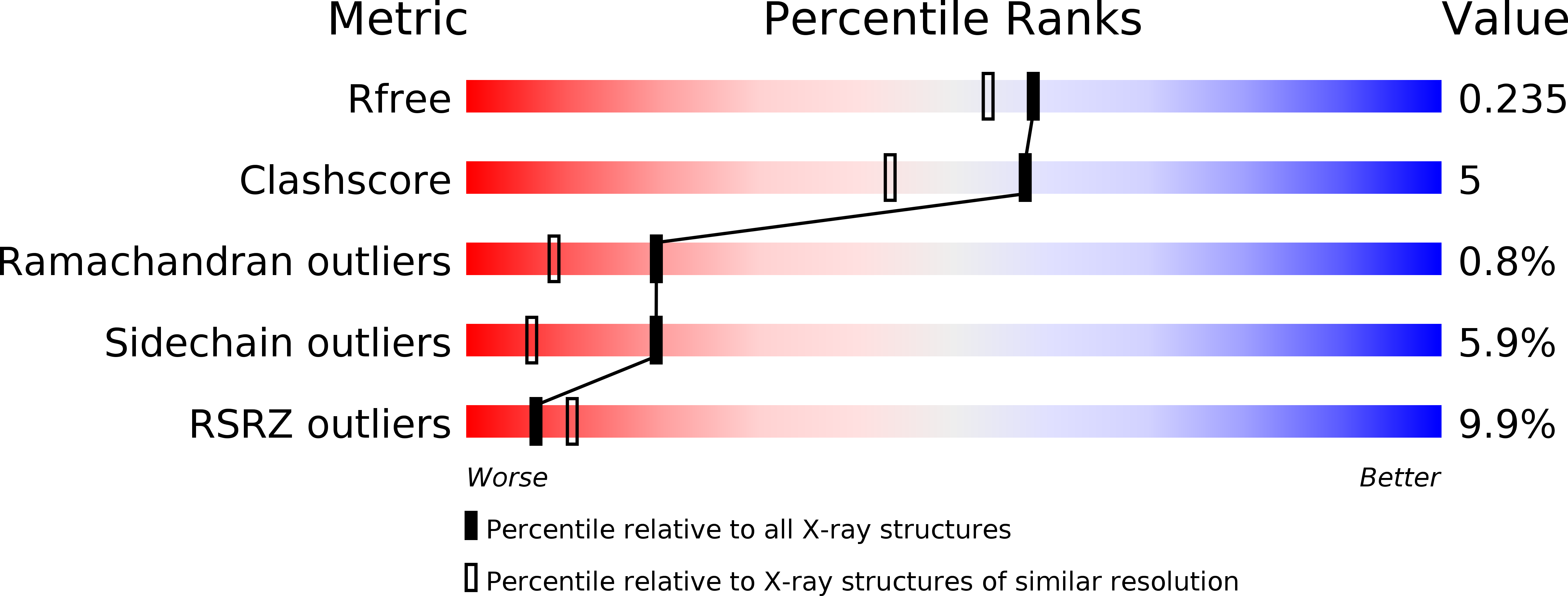
Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Murray, C.W., Berdini, V., Buck, I.M., Carr, M.E., Cleasby, A., Coyle, J.E., Curry, J.E., Day, J.E., Day, P.J., Hearn, K., Iqbal, A., Lee, L.Y., Martins, V., Mortenson, P.N., Munck, J.M., Page, L.W., Patel, S., Roomans, S., Smith, K., Tamanini, E., Saxty, G.(2015) ACS Med Chem Lett 6: 798-803
- PubMed: 26191369
- DOI: https://doi.org/10.1021/acsmedchemlett.5b00143
- Primary Citation of Related Structures:
5BVK, 5BVN, 5BVO, 5BVW - PubMed Abstract:
The DDR1 and DDR2 receptor tyrosine kinases are activated by extracellular collagen and have been implicated in a number of human diseases including cancer. We performed a fragment-based screen against DDR1 and identified fragments that bound either at the hinge or in the back pocket associated with the DFG-out conformation of the kinase. Modeling based on crystal structures of potent kinase inhibitors facilitated the "back-to-front" design of potent DDR1/2 inhibitors that incorporated one of the DFG-out fragments. Further optimization led to low nanomolar, orally bioavailable inhibitors that were selective for DDR1 and DDR2. The inhibitors were shown to potently inhibit DDR2 activity in cells but in contrast to unselective inhibitors such as dasatinib, they did not inhibit proliferation of mutant DDR2 lung SCC cell lines.
Organizational Affiliation:
Astex Pharmaceuticals, 436 Cambridge Science Park, Milton Road, Cambridge CB4 0QA, U.K.