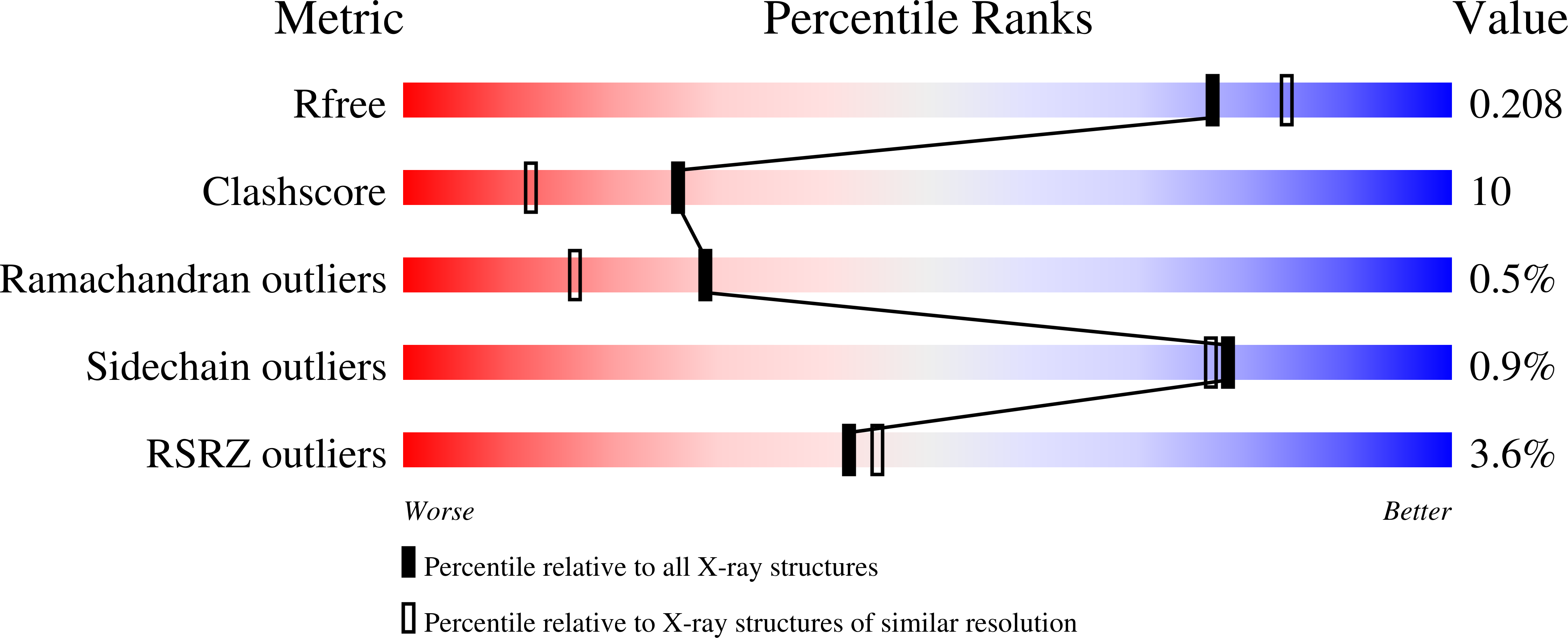
Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
El Gamal, A., Agarwal, V., Diethelm, S., Rahman, I., Schorn, M.A., Sneed, J.M., Louie, G.V., Whalen, K.E., Mincer, T.J., Noel, J.P., Paul, V.J., Moore, B.S.(2016) Proc Natl Acad Sci U S A 113: 3797-3802
- PubMed: 27001835
- DOI: https://doi.org/10.1073/pnas.1519695113
- Primary Citation of Related Structures:
5BUK, 5BUL, 5BVA - PubMed Abstract:
Halogenated pyrroles (halopyrroles) are common chemical moieties found in bioactive bacterial natural products. The halopyrrole moieties of mono- and dihalopyrrole-containing compounds arise from a conserved mechanism in which a proline-derived pyrrolyl group bound to a carrier protein is first halogenated and then elaborated by peptidic or polyketide extensions. This paradigm is broken during the marine pseudoalteromonad bacterial biosynthesis of the coral larval settlement cue tetrabromopyrrole (1), which arises from the substitution of the proline-derived carboxylate by a bromine atom. To understand the molecular basis for decarboxylative bromination in the biosynthesis of 1, we sequenced two Pseudoalteromonas genomes and identified a conserved four-gene locus encoding the enzymes involved in its complete biosynthesis. Through total in vitro reconstitution of the biosynthesis of 1 using purified enzymes and biochemical interrogation of individual biochemical steps, we show that all four bromine atoms in 1 are installed by the action of a single flavin-dependent halogenase: Bmp2. Tetrabromination of the pyrrole induces a thioesterase-mediated offloading reaction from the carrier protein and activates the biosynthetic intermediate for decarboxylation. Insights into the tetrabrominating activity of Bmp2 were obtained from the high-resolution crystal structure of the halogenase contrasted against structurally homologous halogenase Mpy16 that forms only a dihalogenated pyrrole in marinopyrrole biosynthesis. Structure-guided mutagenesis of the proposed substrate-binding pocket of Bmp2 led to a reduction in the degree of halogenation catalyzed. Our study provides a biogenetic basis for the biosynthesis of 1 and sets a firm foundation for querying the biosynthetic potential for the production of 1 in marine (meta)genomes.
Organizational Affiliation:
Center for Oceans and Human Health, Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA 92093;