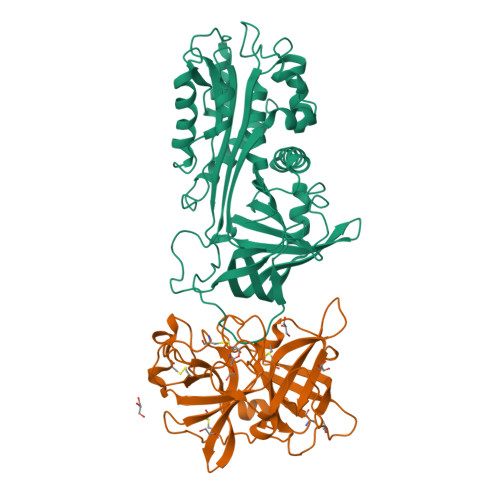
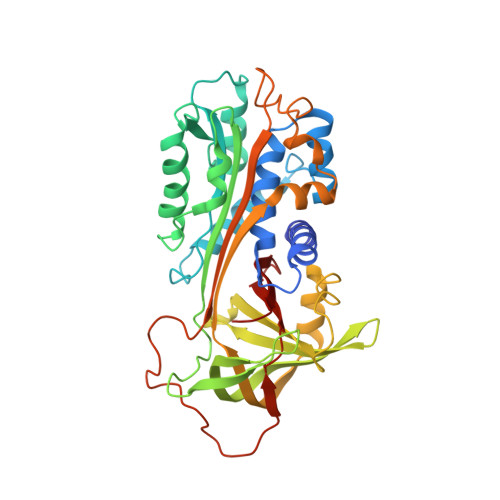
Crystal Structure of the Michaelis Complex between Tissue-type Plasminogen Activator and Plasminogen Activators Inhibitor-1
Gong, L., Liu, M., Zeng, T., Shi, X., Yuan, C., Andreasen, P.A., Huang, M.(2015) J Biological Chem 290: 25795-25804
- PubMed: 26324706
- DOI: https://doi.org/10.1074/jbc.M115.677567
- Primary Citation of Related Structures:
5BRR - PubMed Abstract:
Thrombosis is a leading cause of death worldwide. Recombinant tissue-type plasminogen activator (tPA) is the Food and Drug Administration-approved thrombolytic drug. tPA is rapidly inactivated by endogenous plasminogen activator inhibitor-1 (PAI-1). Engineering on tPA to reduce its inhibition by PAI-1 without compromising its thrombolytic effect is a continuous effort. Precise details, with atomic resolution, of the molecular interactions between tPA and PAI-1 remain unknown despite previous extensive studies. Here, we report the crystal structure of the tPA·PAI-1 Michaelis complex, which shows significant differences from the structure of its urokinase-type plasminogen activator analogue, the uPA·PAI-1 Michaelis complex. The PAI-1 reactive center loop adopts a unique kinked conformation. The structure provides detailed interactions between tPA 37- and 60-loops with PAI-1. On the tPA side, the S2 and S1β pockets open up to accommodate PAI-1. This study provides structural basis to understand the specificity of PAI-1 and to design newer generation of thrombolytic agents with reduced PAI-1 inactivation.
Organizational Affiliation:
From the State Key Laboratory of Structural Chemistry and Danish-Chinese Centre for Proteases and Cancer, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fuzhou, 350002 Fujian, China, the University of Chinese Academy of Sciences, Beijing, 100049, China, and.