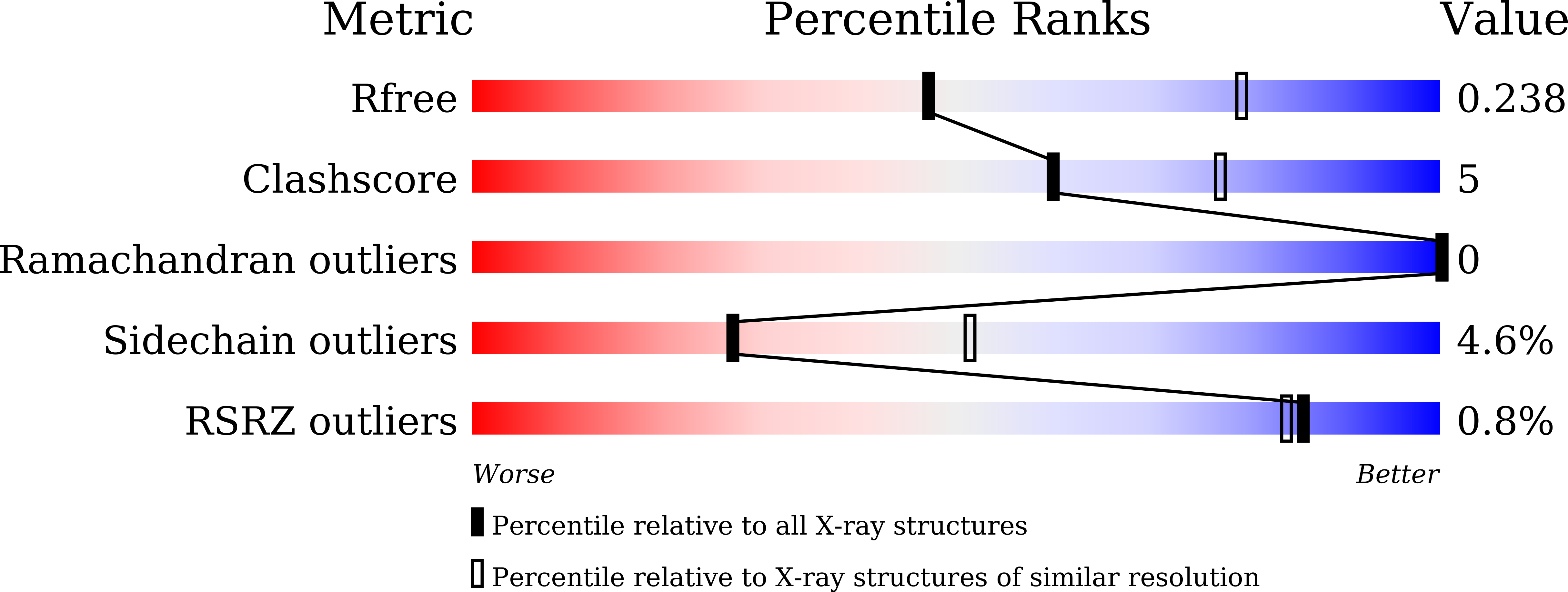
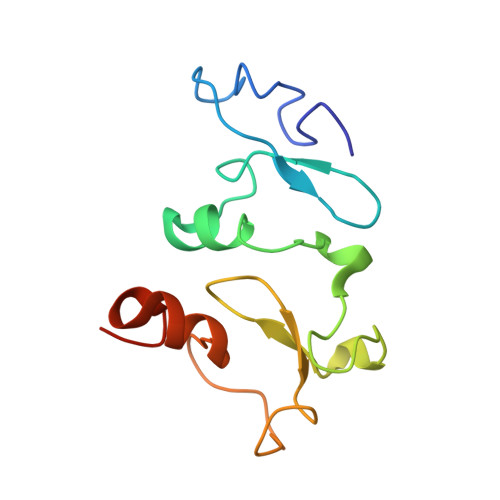
Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2
Xiong, X., Panchenko, T., Yang, S., Zhao, S., Yan, P., Zhang, W., Xie, W., Li, Y., Zhao, Y., Allis, C.D., Li, H.(2016) Nat Chem Biol 12: 1111-1118
- PubMed: 27775714
- DOI: https://doi.org/10.1038/nchembio.2218
- Primary Citation of Related Structures:
5B75, 5B76, 5B77, 5B78, 5B79 - PubMed Abstract:
Recognition of histone covalent modifications by 'reader' modules constitutes a major mechanism for epigenetic regulation. A recent upsurge of newly discovered histone lysine acylations, such as crotonylation (Kcr), butyrylation (Kbu), and propionylation (Kpr), greatly expands the coding potential of histone lysine modifications. Here we demonstrate that the histone acetylation-binding double PHD finger (DPF) domains of human MOZ (also known as KAT6A) and DPF2 (also known as BAF45d) accommodate a wide range of histone lysine acylations with the strongest preference for Kcr. Crystal structures of the DPF domain of MOZ in complex with H3K14cr, H3K14bu, and H3K14pr peptides reveal that these non-acetyl acylations are anchored in a hydrophobic 'dead-end' pocket with selectivity for crotonylation arising from intimate encapsulation and an amide-sensing hydrogen bonding network. Immunofluorescence and chromatin immunoprecipitation (ChIP)-quantitative PCR (qPCR) showed that MOZ and H3K14cr colocalize in a DPF-dependent manner. Our studies call attention to a new regulatory mechanism centered on histone crotonylation readout by DPF family members.
Organizational Affiliation:
MOE Key Laboratory of Protein Sciences, Beijing Advanced Innovation Center for Structural Biology, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing, China.