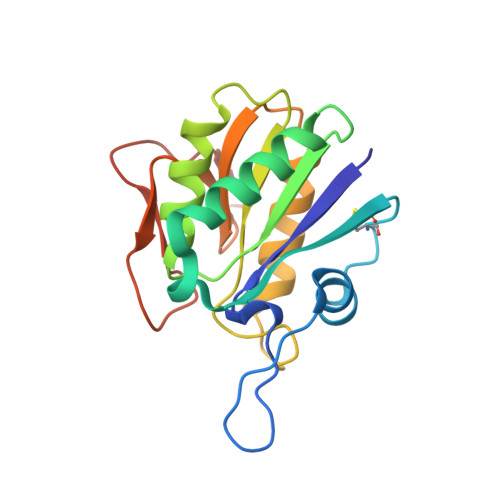
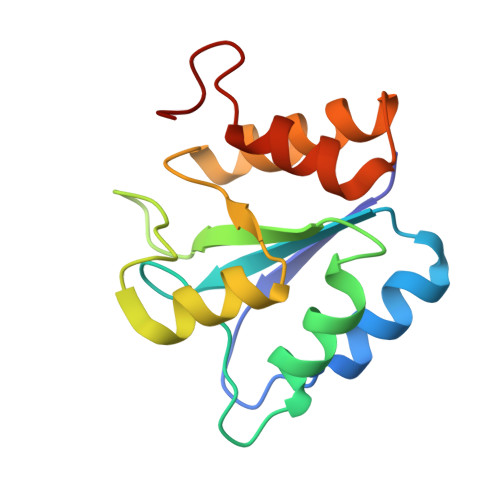
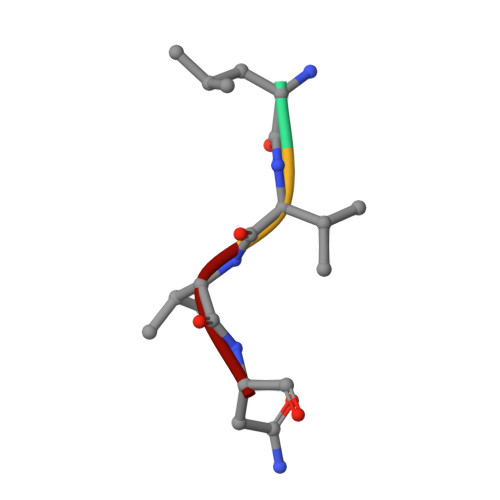
Heat induces end to end repetitive association in P. furiosus L-asparaginase which enables its thermophilic property.
Sharma, P., Tomar, R., Yadav, S.S., Badmalia, M.D., Nath, S.K., Kundu, B.(2020) Sci Rep 10: 21702-21702
- PubMed: 33303914
- DOI: https://doi.org/10.1038/s41598-020-78877-z
- Primary Citation of Related Structures:
5B5U, 5B74, 5CBP - PubMed Abstract:
It remains undeciphered how thermophilic enzymes display enhanced stability at elevated temperatures. Taking L-asparaginase from P. furiosus (PfA) as an example, we combined scattering shapes deduced from small-angle X-ray scattering (SAXS) data at increased temperatures with symmetry mates from crystallographic structures to find that heating caused end-to-end association. The small contact point of self-binding appeared to be enabled by a terminal short β-strand in N-terminal domain, Leu 179 -Val-Val-Asn 182 (LVVN). Interestingly, deletion of this strand led to a defunct enzyme, whereas suplementation of the peptide LVVN to the defunct enzyme restored structural frameworkwith mesophile-type functionality. Crystal structure of the peptide-bound defunct enzyme showed that one peptide ispresent in the same coordinates as in original enzyme, explaining gain-of lost function. A second peptide was seen bound to the protein at a different location suggesting its possible role in substrate-free molecular-association. Overall, we show that the heating induced self-assembly of native shapes of PfA led to an apparent super-stable assembly.
Organizational Affiliation:
CSIR-Institute of Microbial Technology, Sec 39 A, Chandigarh, 160036, India.