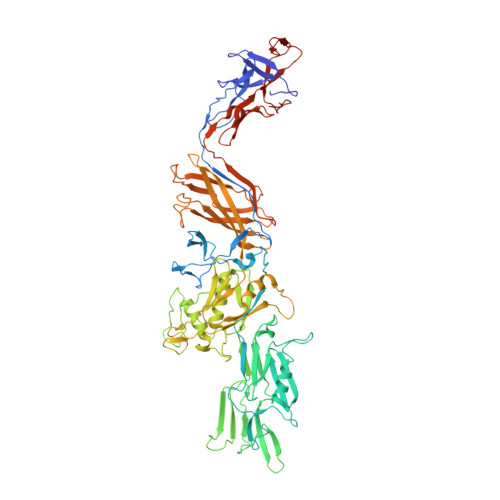
Structural insights into bacterial flagellar hooks similarities and specificities.
Yoon, Y.-H., Barker, C.S., Bulieris, P.V., Matsunami, H., Samatey, F.A.(2016) Sci Rep 6: 35552-35552
- PubMed: 27759043
- DOI: https://doi.org/10.1038/srep35552
- Primary Citation of Related Structures:
5AY6, 5AZ4 - PubMed Abstract:
Across bacteria, the protein that makes the flagellar hook, FlgE, has a high variability in amino acid residue composition and sequence length. We hereby present the structure of two fragments of FlgE protein from Campylobacter jejuni and from Caulobacter crescentus, which were obtained by X-ray crystallography, and a high-resolution model of the hook from Caulobacter. By comparing these new structures of FlgE proteins, we show that bacterial hook can be divided in two distinct parts. The first part comprises domains that are found in all FlgE proteins and that will make the basic structure of the hook that is common to all flagellated bacteria. The second part, hyper-variable both in size and structure, will be bacteria dependent. To have a better understanding of the C. jejuni hook, we show that a special strain of Salmonella enterica, which was designed to encode a gene of flgE that has the extra domains found in FlgE from C. jejuni, is fully motile. It seems that no matter the size of the hook protein, the hook will always have a structure made of 11 protofilaments.
Organizational Affiliation:
Trans-Membrane Trafficking Unit, Okinawa Institute of Science and Technology Graduate University, 1919-1, Tancha, Onna, Kunigami, Okinawa 904-0495, Japan.