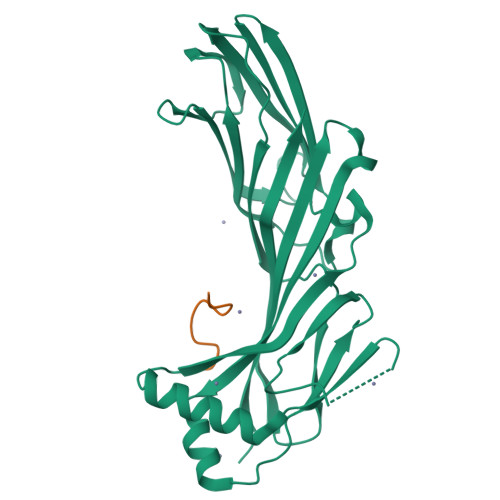
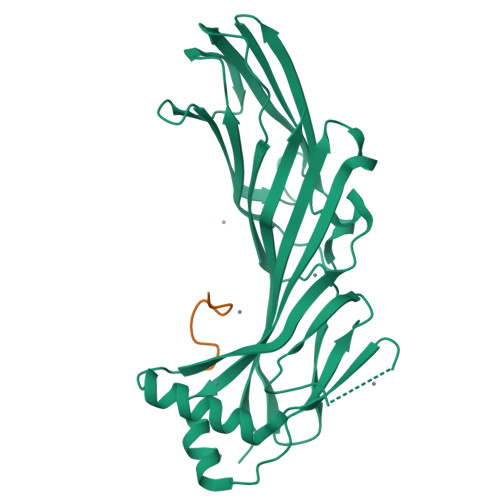
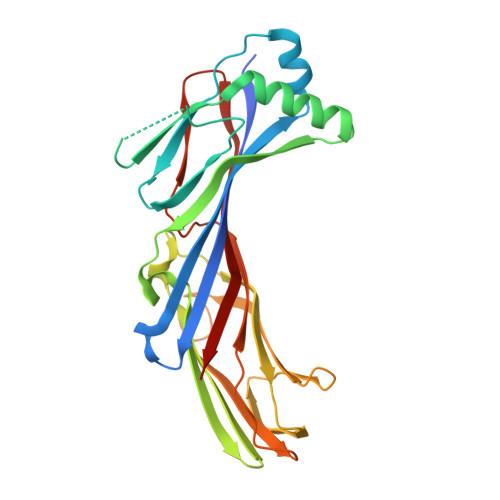
Structural basis for the recognition of two consecutive mutually interacting DPF motifs by the SGIP1 mu homology domain.
Shimada, A., Yamaguchi, A., Kohda, D.(2016) Sci Rep 6: 19565-19565
- PubMed: 26822536
- DOI: https://doi.org/10.1038/srep19565
- Primary Citation of Related Structures:
5AWR, 5AWS, 5AWT, 5AWU - PubMed Abstract:
FCHo1, FCHo2, and SGIP1 are key regulators of clathrin-mediated endocytosis. Their μ homology domains (μHDs) interact with the C-terminal region of an endocytic scaffold protein, Eps15, containing fifteen Asp-Pro-Phe (DPF) motifs. Here, we show that the high-affinity μHD-binding site in Eps15 is a region encompassing six consecutive DPF motifs, while the minimal μHD-binding unit is two consecutive DPF motifs. We present the crystal structures of the SGIP1 μHD in complex with peptides containing two DPF motifs. The peptides bind to a novel ligand-binding site of the μHD, which is distinct from those of other distantly related μHD-containing proteins. The two DPF motifs, which adopt three-dimensional structures stabilized by sequence-specific intramotif and intermotif interactions, are extensively recognized by the μHD and are both required for binding. Thus, consecutive and singly scattered DPF motifs play distinct roles in μHD binding.
Organizational Affiliation:
Division of Structural Biology, Medical Institute of Bioregulation, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.