Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
Ferrandi, E.E., Sayer, C., Isupov, M.N., Annovazzi, C., Marchesi, C., Iacobone, G., Peng, X., Bonch-Osmolovskaya, E., Wohlgemuth, R., Littlechild, J.A., Montia, D.(2015) FEBS J 282: 2879
- PubMed: 26032250
- DOI: https://doi.org/10.1111/febs.13328
- Primary Citation of Related Structures:
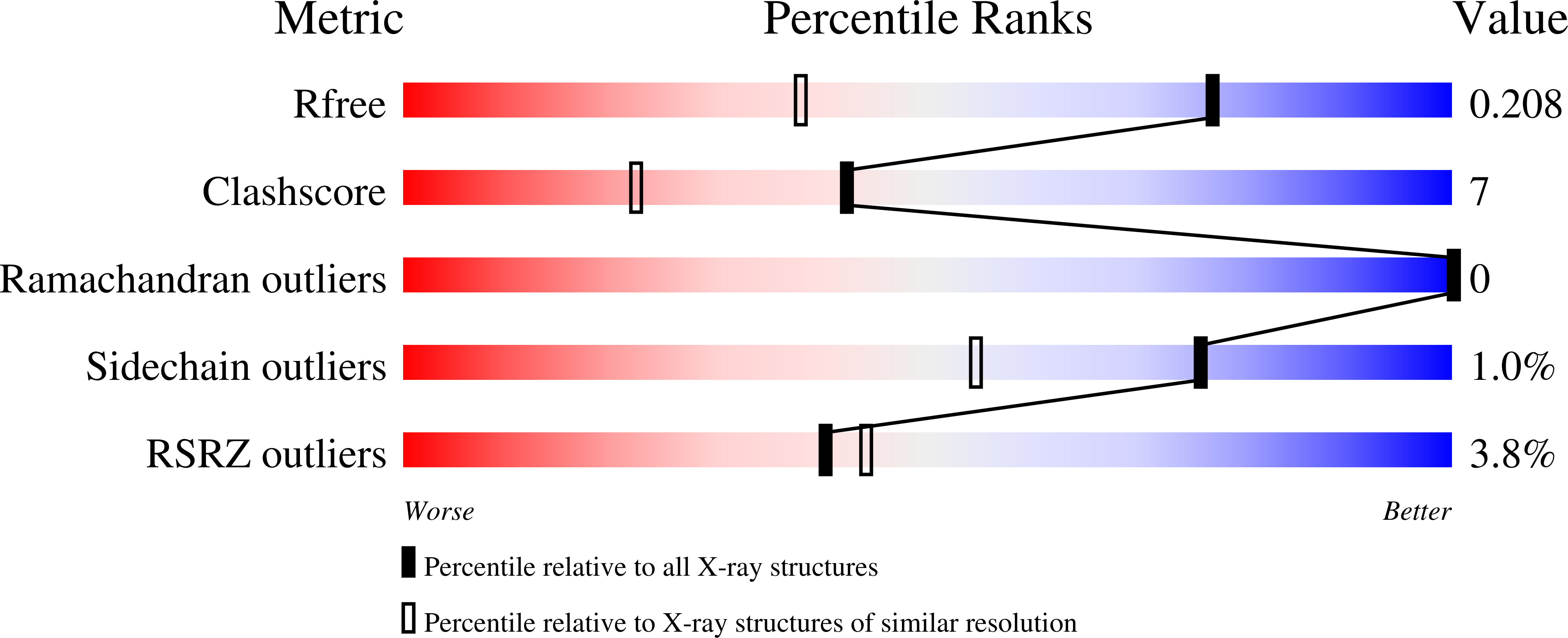
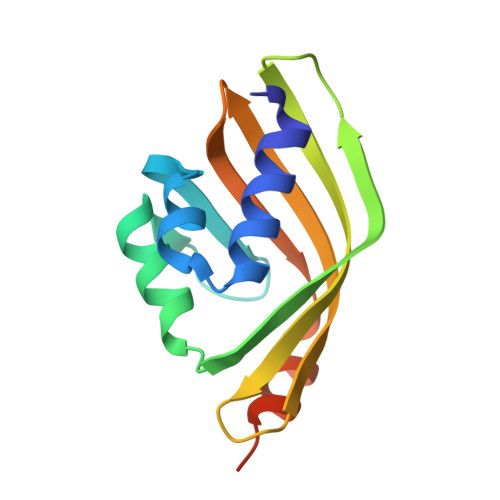
5AIF, 5AIG, 5AIH, 5AII - PubMed Abstract:
The epoxide hydrolases (EHs) represent an attractive option for the synthesis of chiral epoxides and 1,2-diols which are valuable building blocks for the synthesis of several pharmaceutical compounds. A metagenomic approach has been used to identify two new members of the atypical EH limonene-1,2-epoxide hydrolase (LEH) family of enzymes. These two LEHs (Tomsk-LEH and CH55-LEH) show EH activities towards different epoxide substrates, differing in most cases from those previously identified for Rhodococcus erythropolis (Re-LEH) in terms of stereoselectivity. Tomsk-LEH and CH55-LEH, both from thermophilic sources, have higher optimal temperatures and apparent melting temperatures than Re-LEH. The new LEH enzymes have been crystallized and their structures solved to high resolution in the native form and in complex with the inhibitor valpromide for Tomsk-LEH and poly(ethylene glycol) for CH55-LEH. The structural analysis has provided insights into the LEH mechanism, substrate specificity and stereoselectivity of these new LEH enzymes, which has been supported by mutagenesis studies.
Organizational Affiliation:
Istituto di Chimica del Riconoscimento Molecolare, C.N.R., Milano, Italy.