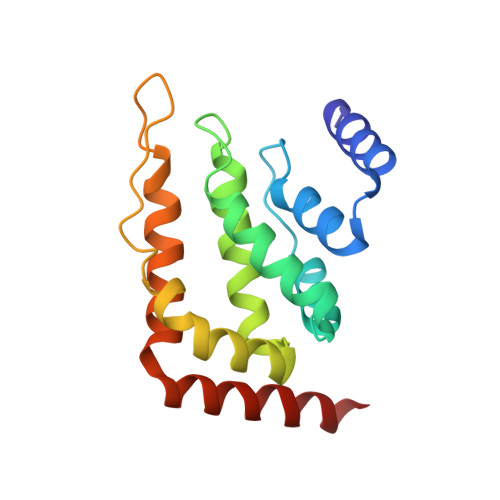
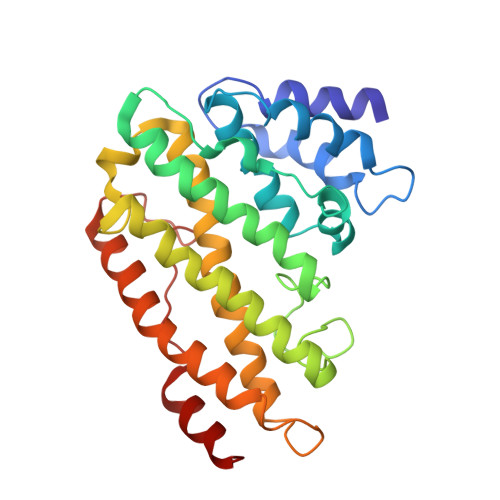
An Organized Co-Assembly of Clathrin Adaptors is Essential for Endocytosis.
Skruzny, M., Desfosses, A., Prinz, S., Dodonova, S.O., Gieras, A., Uetrecht, C., Jakobi, A.J., Abella, M., Hagen, W.J.H., Schulz, J., Meijers, R., Rybin, V., Briggs, J.A.G., Sachse, C., Kaksonen, M.(2015) Dev Cell 33: 150
- PubMed: 25898165
- DOI: https://doi.org/10.1016/j.devcel.2015.02.023
- Primary Citation of Related Structures:
5AHV - PubMed Abstract:
Clathrin-mediated endocytosis, the main trafficking route from the plasma membrane to the cytoplasm, is critical to many fundamental cellular processes. Clathrin, coupled to the membrane by adaptor proteins, is thought to play a major structural role in endocytosis by self-assembling into a cage-like lattice around the forming vesicle. Although clathrin adaptors are essential for endocytosis, little is known about their structural role in this process. Here we show that the membrane-binding domains of two conserved clathrin adaptors, Sla2 and Ent1, co-assemble in a PI(4,5)P2-dependent manner to form organized lattices on membranes. We determined the structure of the co-assembled lattice by electron cryo-microscopy and designed mutations that specifically impair the lattice formation in vitro. We show that these mutations block endocytosis in vivo. We suggest that clathrin adaptors not only link the polymerized clathrin to the membrane but also form an oligomeric structure, which is essential for membrane remodeling during endocytosis.
- Cell Biology and Biophysics Unit, European Molecular Biology Laboratory (EMBL), 69117 Heidelberg, Germany.
Organizational Affiliation: