Biophysical Studies of the Induced Dimerization of Human Vegf R Receptor 1 Binding Domain by Divalent Metals Competing with Vegf-A
Gaucher, J.-F., Reille-Seroussi, M., Gagey-Eilstein, N., Broussy, S., Coric, P., Seijo, B., Lascombe, M.-B., Gautier, B., Liu, W.-Q., Huguenot, F., Inguimbert, N., Bouaziz, S., Vidal, M., Broutin, I.(2016) PLoS One 11: 67755
- PubMed: 27942001
- DOI: https://doi.org/10.1371/journal.pone.0167755
- Primary Citation of Related Structures:
4CKV, 4CL7, 5ABD - PubMed Abstract:
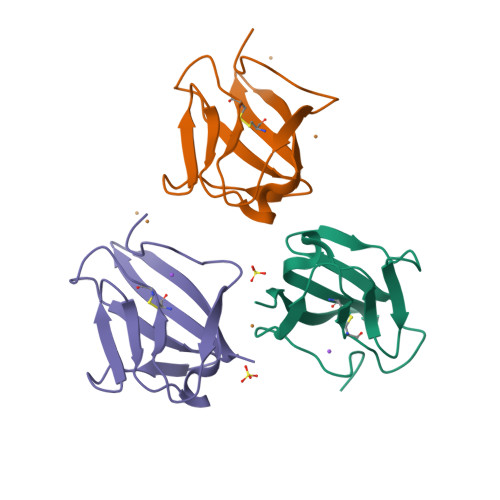
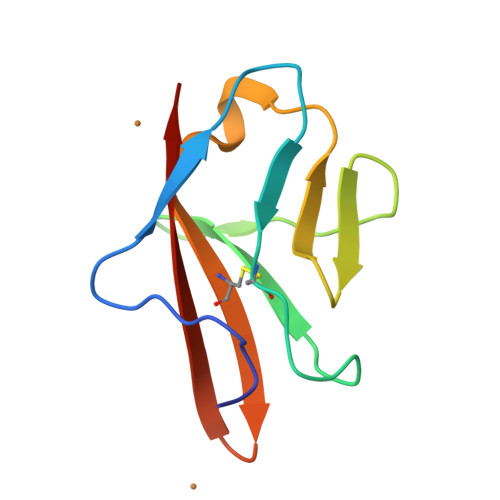
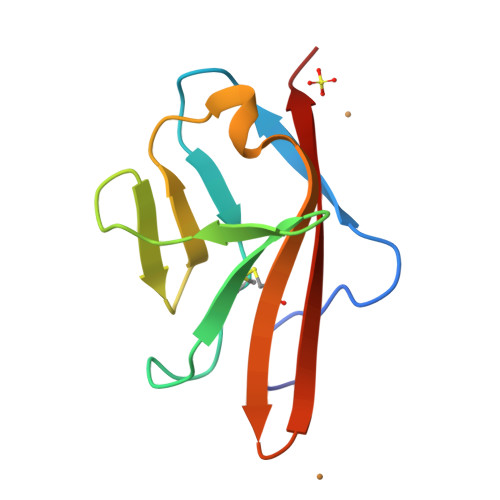
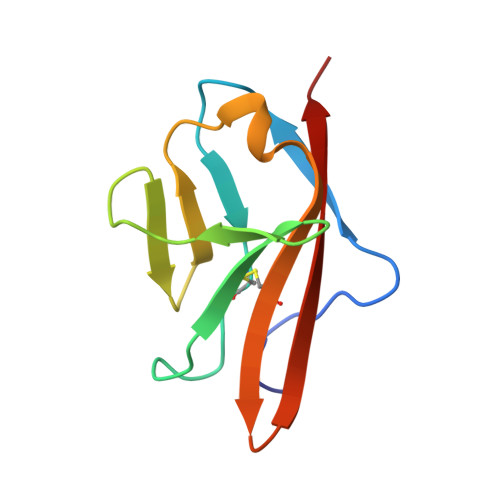
Angiogenesis is tightly regulated through the binding of vascular endothelial growth factors (VEGFs) to their receptors (VEGFRs). In this context, we showed that human VEGFR1 domain 2 crystallizes in the presence of Zn2+, Co2+ or Cu2+ as a dimer that forms via metal-ion interactions and interlocked hydrophobic surfaces. SAXS, NMR and size exclusion chromatography analyses confirm the formation of this dimer in solution in the presence of Co2+, Cd2+ or Cu2+. Since the metal-induced dimerization masks the VEGFs binding surface, we investigated the ability of metal ions to displace the VEGF-A binding to hVEGFR1: using a competition assay, we evidenced that the metals displaced the VEGF-A binding to hVEGFR1 extracellular domain binding at micromolar level.
Organizational Affiliation:
UMR 8015 CNRS - Université Paris Descartes, Faculté de Pharmacie, Sorbonne Paris Cité, Paris, France.