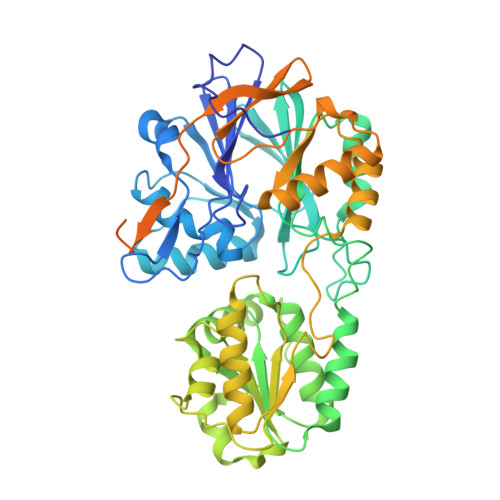
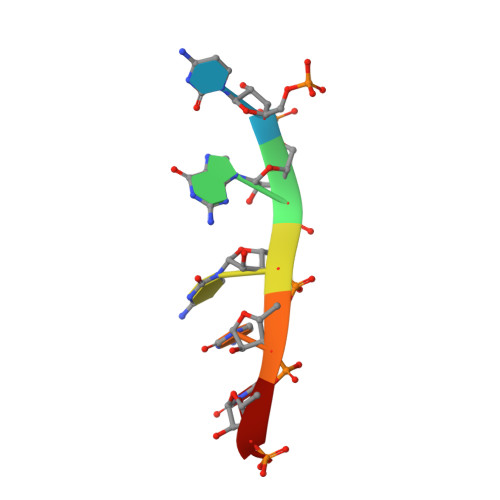
Linkage of Catalysis and 5' End Recognition in Ribonuclease Rnase J
Pei, X.Y., Bralley, P., Jones, G.H., Luisi, B.F.(2015) Nucleic Acids Res 43: 8066
- PubMed: 26253740
- DOI: https://doi.org/10.1093/nar/gkv732
- Primary Citation of Related Structures:
5A0T, 5A0V - PubMed Abstract:
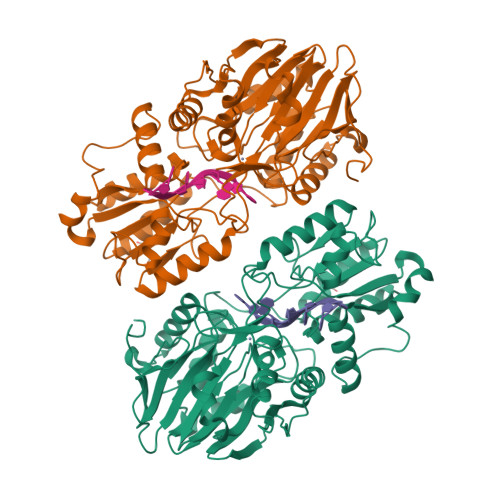
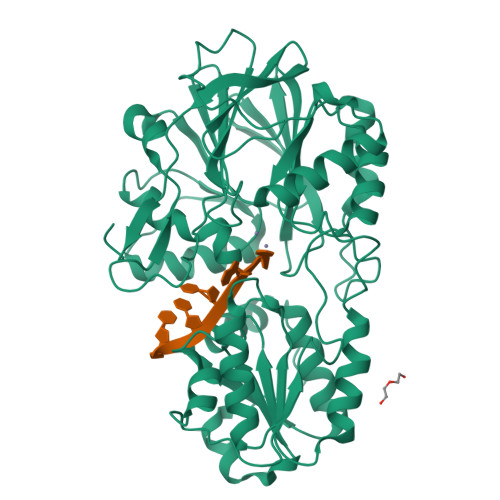
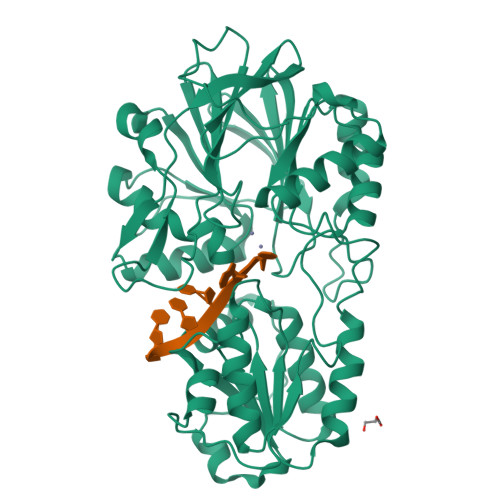
In diverse bacterial species, the turnover and processing of many RNAs is mediated by the ribonuclease RNase J, a member of the widely occurring metallo-β-lactamase enzyme family. We present crystal structures of Streptomyces coelicolor RNase J with bound RNA in pre- and post-cleavage states, at 2.27 Å and 2.80 Å resolution, respectively. These structures reveal snapshots of the enzyme cleaving substrate directionally and sequentially from the 5' terminus. In the pre-cleavage state, a water molecule is coordinated to a zinc ion pair in the active site but is imperfectly oriented to launch a nucleophilic attack on the phosphate backbone. A conformational switch is envisaged that enables the in-line positioning of the attacking water and may be facilitated by magnesium ions. Adjacent to the scissile bond, four bases are stacked in a tightly sandwiching pocket, and mutagenesis results indicate that this organization helps to drive processive exo-ribonucleolytic cleavage. Like its numerous homologues, S. coelicolor RNase J can also cleave some RNA internally, and the structural data suggest how the preference for exo- versus endo-cleavage mode is linked with recognition of the chemical status of the substrate's 5' end.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, Tennis Court Road, Cambridge CB2 1GA, UK.