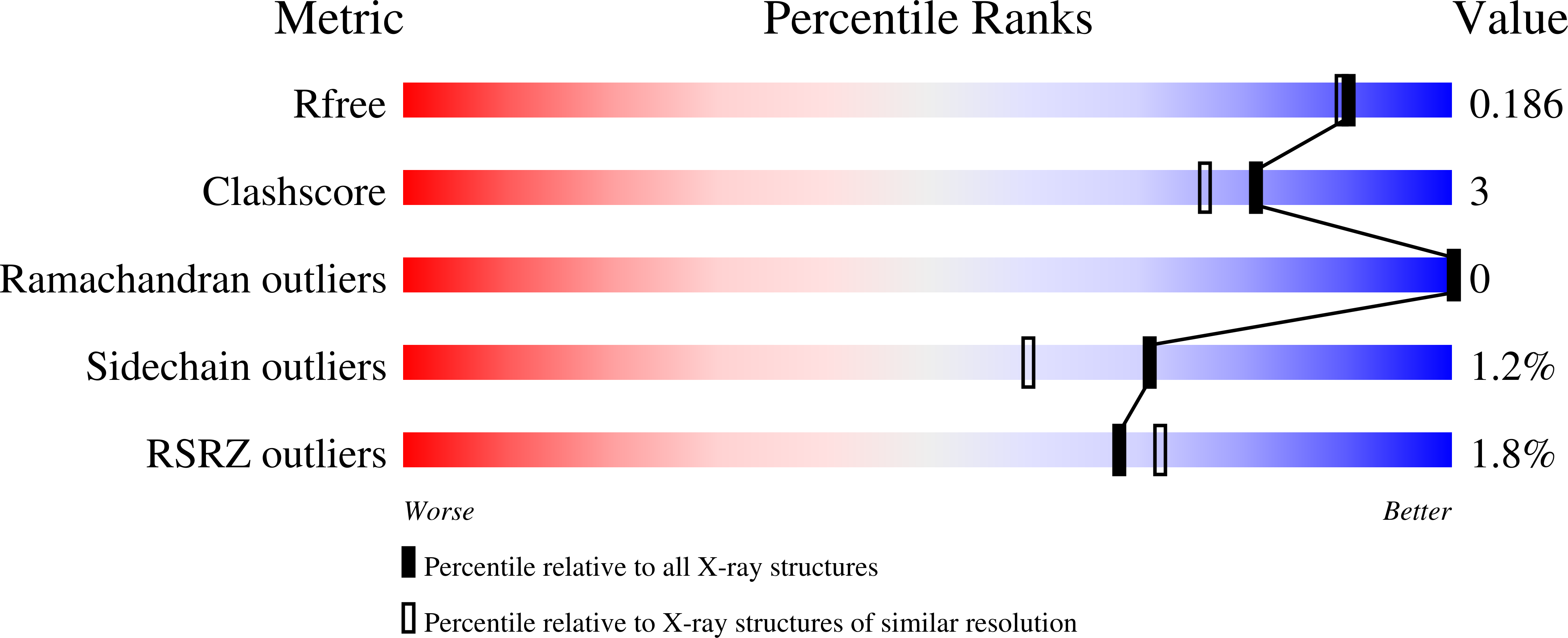
Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Taberman, H., Andberg, M., Koivula, A., Hakulinen, N., Penttila, M., Rouvinen, J., Parkkinen, T.(2015) Biochem J 472: 297
- PubMed: 26438878
- DOI: https://doi.org/10.1042/BJ20150681
- Primary Citation of Related Structures:
5A02, 5A03, 5A04, 5A05, 5A06 - PubMed Abstract:
Aldose-aldose oxidoreductase (Cc AAOR) is a recently characterized enzyme from the bacterial strain Caulobacter crescentus CB15 belonging to the glucose-fructose oxidoreductase/inositol dehydrogenase/rhizopine catabolism protein (Gfo/Idh/MocA) family. Cc AAOR catalyses the oxidation and reduction of a panel of aldose monosaccharides using a tightly bound NADP(H) cofactor that is regenerated in the catalytic cycle. Furthermore, Cc AAOR can also oxidize 1,4-linked oligosaccharides. In the present study, we present novel crystal structures of the dimeric Cc AAOR in complex with the cofactor and glycerol, D-xylose, D-glucose, maltotriose and D-sorbitol determined to resolutions of 2.0, 1.8, 1.7, 1.9 and 1.8 Å (1 Å=0.1 nm), respectively. These complex structures allowed for a detailed analysis of the ligand-binding interactions. The structures showed that the C1 carbon of a substrate, which is either reduced or oxidized, is close to the reactive C4 carbon of the nicotinamide ring of NADP(H). In addition, the O1 hydroxy group of the substrate, which is either protonated or deprotonated, is unexpectedly close to both Lys(104) and Tyr(189), which may both act as a proton donor or acceptor. This led us to hypothesize that this intriguing feature could be beneficial for Cc AAOR to catalyse the reduction of a linear form of a monosaccharide substrate and the oxidation of a pyranose form of the same substrate in a reaction cycle, during which the bound cofactor is regenerated.
Organizational Affiliation:
Department of Chemistry, University of Eastern Finland, PO Box 111, FI-80101 Joensuu, Finland.