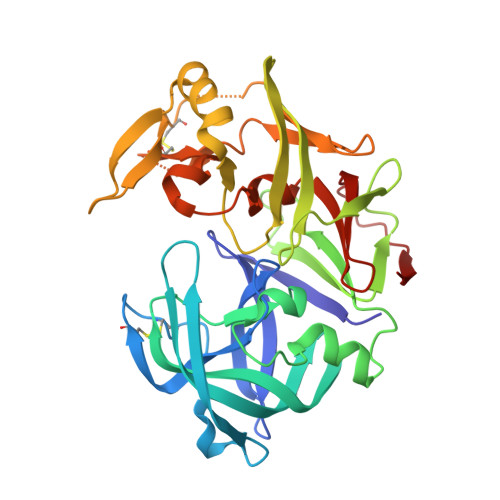
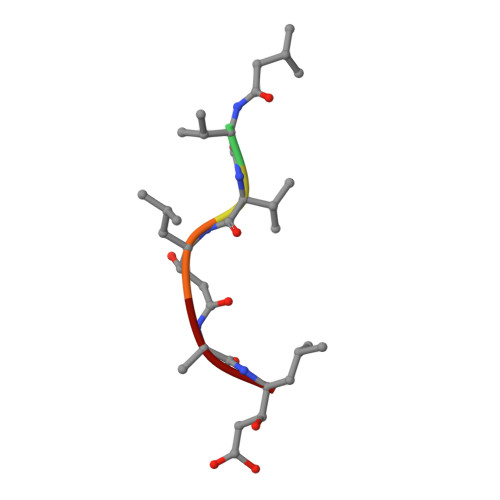
Atomic resolution crystal structure of Sapp2p, a secreted aspartic protease from Candida parapsilosis.
Dostal, J., Pecina, A., Hruskova-Heidingsfeldova, O., Mareckova, L., Pichova, I., Rezacova, P., Lepsik, M., Brynda, J.(2015) Acta Crystallogr D Biol Crystallogr 71: 2494-2504
- PubMed: 26627656
- DOI: https://doi.org/10.1107/S1399004715019392
- Primary Citation of Related Structures:
4Y9W, 4YBF - PubMed Abstract:
The virulence of the Candida pathogens is enhanced by the production of secreted aspartic proteases, which therefore represent possible targets for drug design. Here, the crystal structure of the secreted aspartic protease Sapp2p from Candida parapsilosis was determined. Sapp2p was isolated from its natural source and crystallized in complex with pepstatin A, a classical aspartic protease inhibitor. The atomic resolution of 0.83 Å allowed the protonation states of the active-site residues to be inferred. A detailed comparison of the structure of Sapp2p with the structure of Sapp1p, the most abundant C. parapsilosis secreted aspartic protease, was performed. The analysis, which included advanced quantum-chemical interaction-energy calculations, uncovered molecular details that allowed the experimentally observed equipotent inhibition of both isoenzymes by pepstatin A to be rationalized.
- Institute of Organic Chemistry and Biochemistry (IOCB), Academy of Sciences of the Czech Republic, Flemingovo náměstí 2, 166 10 Prague 6, Czech Republic.
Organizational Affiliation: