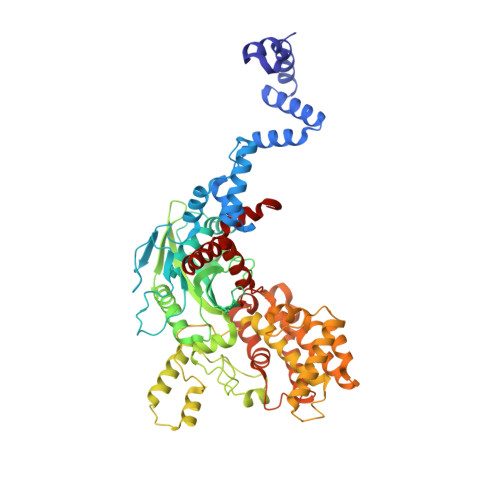
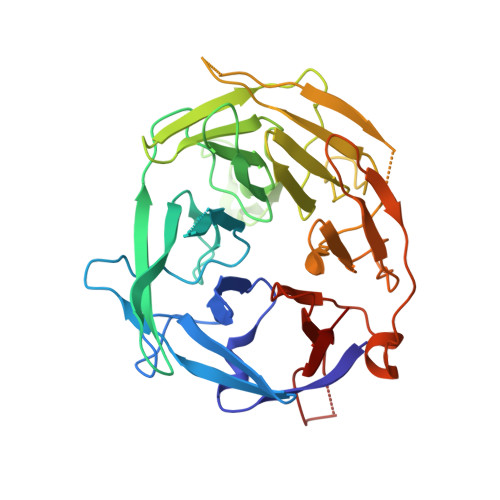
Crystal structure of the V(D)J recombinase RAG1-RAG2.
Kim, M.S., Lapkouski, M., Yang, W., Gellert, M.(2015) Nature 518: 507-511
- PubMed: 25707801
- DOI: https://doi.org/10.1038/nature14174
- Primary Citation of Related Structures:
4WWX - PubMed Abstract:
V(D)J recombination in the vertebrate immune system generates a highly diverse population of immunoglobulins and T-cell receptors by combinatorial joining of segments of coding DNA. The RAG1-RAG2 protein complex initiates this site-specific recombination by cutting DNA at specific sites flanking the coding segments. Here we report the crystal structure of the mouse RAG1-RAG2 complex at 3.2 Å resolution. The 230-kilodalton RAG1-RAG2 heterotetramer is 'Y-shaped', with the amino-terminal domains of the two RAG1 chains forming an intertwined stalk. Each RAG1-RAG2 heterodimer composes one arm of the 'Y', with the active site in the middle and RAG2 at its tip. The RAG1-RAG2 structure rationalizes more than 60 mutations identified in immunodeficient patients, as well as a large body of genetic and biochemical data. The architectural similarity between RAG1 and the hairpin-forming transposases Hermes and Tn5 suggests the evolutionary conservation of these DNA rearrangements.
- Laboratory of Molecular Biology, NIDDK, NIH, Bethesda, Maryland 20892, USA.
Organizational Affiliation: