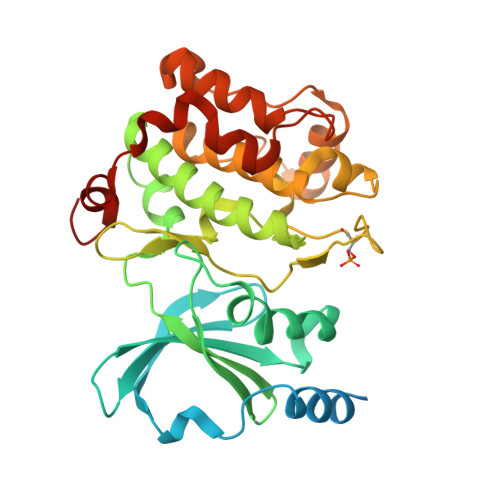
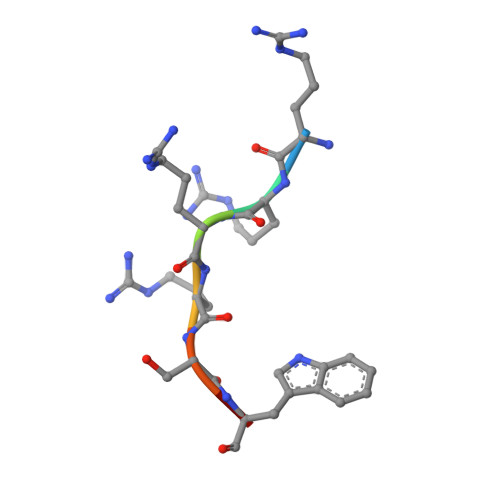
Identification of a major determinant for serine-threonine kinase phosphoacceptor specificity.
Chen, C., Ha, B.H., Thevenin, A.F., Lou, H.J., Zhang, R., Yip, K.Y., Peterson, J.R., Gerstein, M., Kim, P.M., Filippakopoulos, P., Knapp, S., Boggon, T.J., Turk, B.E.(2014) Mol Cell 53: 140-147
- PubMed: 24374310
- DOI: https://doi.org/10.1016/j.molcel.2013.11.013
- Primary Citation of Related Structures:
4JDH, 4JDI, 4JDJ, 4JDK - PubMed Abstract:
Eukaryotic protein kinases are generally classified as being either tyrosine or serine-threonine specific. Though not evident from inspection of their primary sequences, many serine-threonine kinases display a significant preference for serine or threonine as the phosphoacceptor residue. Here we show that a residue located in the kinase activation segment, which we term the "DFG+1" residue, acts as a major determinant for serine-threonine phosphorylation site specificity. Mutation of this residue was sufficient to switch the phosphorylation site preference for multiple kinases, including the serine-specific kinase PAK4 and the threonine-specific kinase MST4. Kinetic analysis of peptide substrate phosphorylation and crystal structures of PAK4-peptide complexes suggested that phosphoacceptor residue preference is not mediated by stronger binding of the favored substrate. Rather, favored kinase-phosphoacceptor combinations likely promote a conformation optimal for catalysis. Understanding the rules governing kinase phosphoacceptor preference allows kinases to be classified as serine or threonine specific based on their sequence.
- Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06520, USA.
Organizational Affiliation: